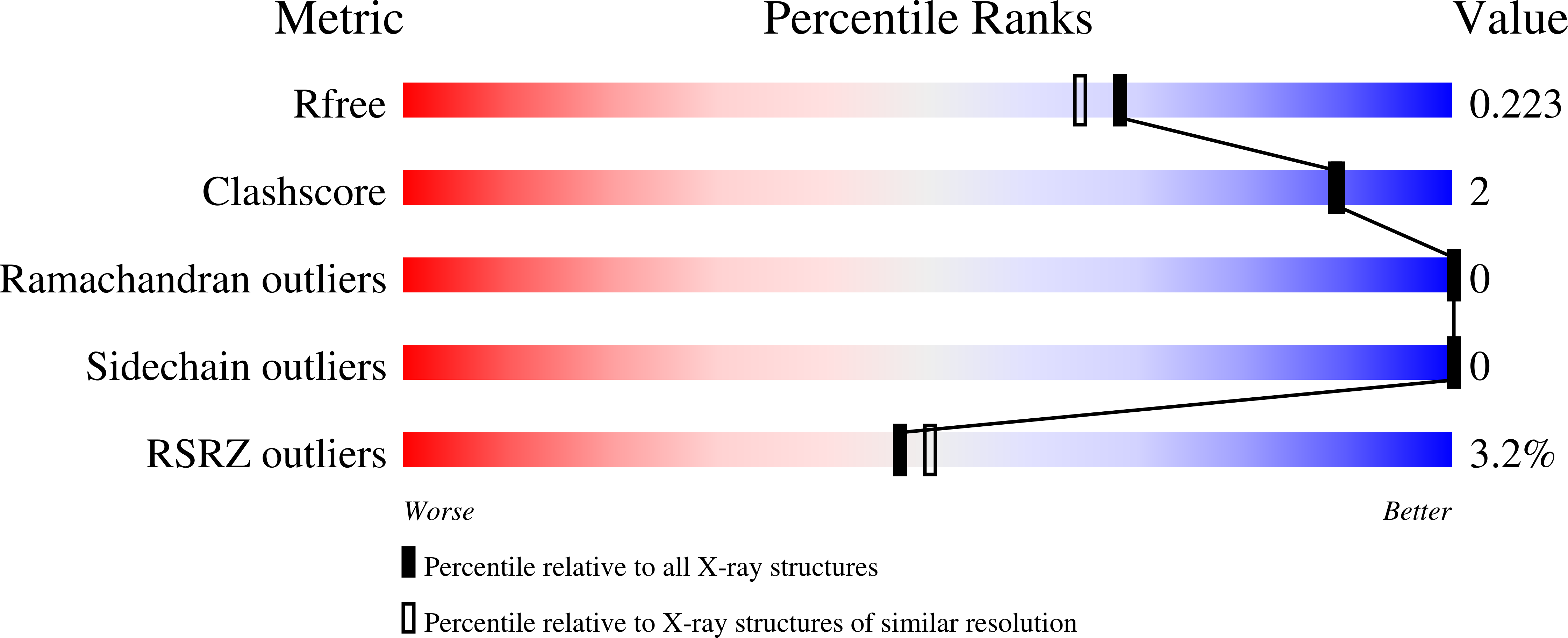
1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
Minasov, G., Wawrzak, Z., Skarina, T., McChesney, C., Grimshaw, S., Sandoval, J., Satchell, K.J.F., Savchenko, A., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.