A bacterial immunomodulatory protein with lipocalin-like domains facilitates host-bacteria mutualism in larval zebrafish.
Rolig, A.S., Sweeney, E.G., Kaye, L.E., DeSantis, M.D., Perkins, A., Banse, A.V., Hamilton, M.K., Guillemin, K.(2018) Elife 7
- PubMed: 30398151
- DOI: https://doi.org/10.7554/eLife.37172
- Primary Citation of Related Structures:
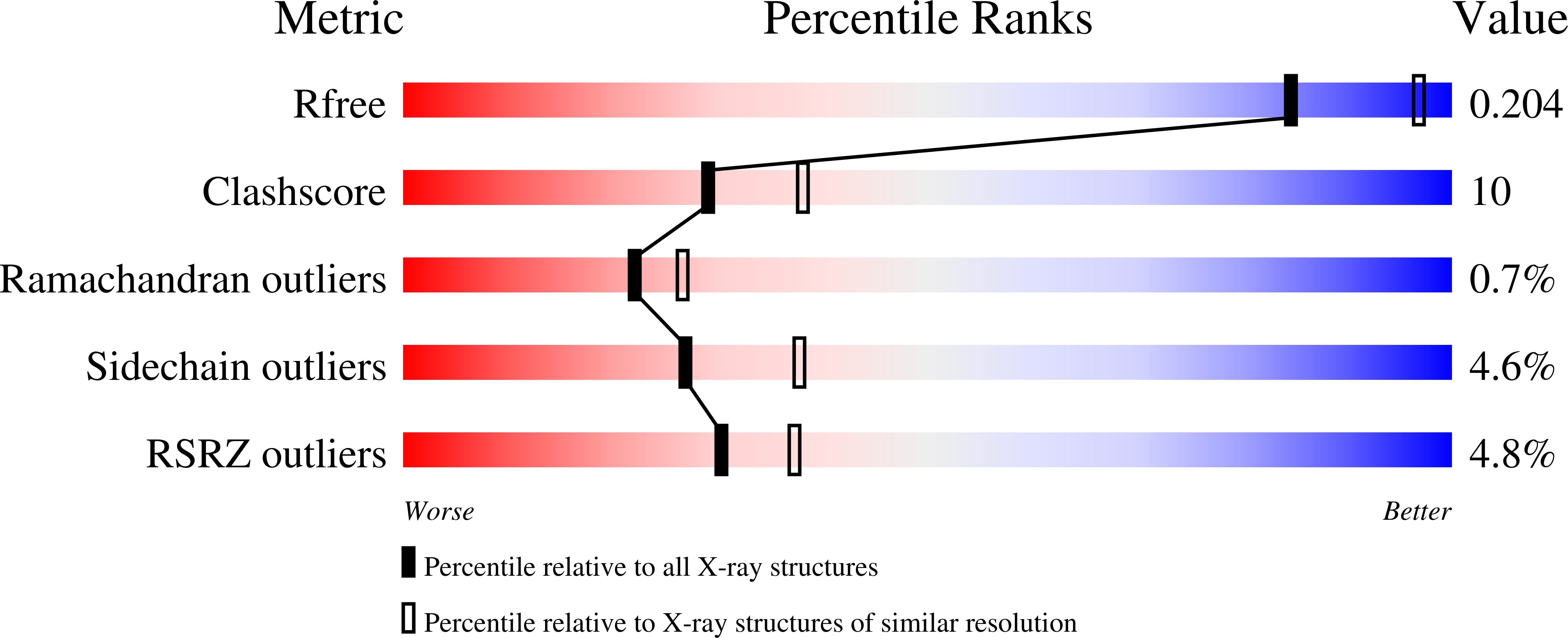
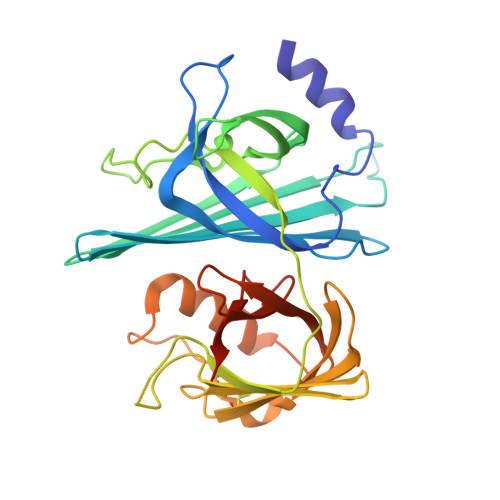
6B7L - PubMed Abstract:
Stable mutualism between a host and its resident bacteria requires a moderated immune response to control bacterial population size without eliciting excessive inflammation that could harm both partners. Little is known about the specific molecular mechanisms utilized by bacterial mutualists to temper their hosts' responses and protect themselves from aggressive immune attack. Using a gnotobiotic larval zebrafish model, we identified an Aeromonas secreted immunomodulatory protein, AimA. AimA is required during colonization to prevent intestinal inflammation that simultaneously compromises both bacterial and host survival. Administration of exogenous AimA prevents excessive intestinal neutrophil accumulation and protects against septic shock in models of both bacterially and chemically induced intestinal inflammation. We determined the molecular structure of AimA, which revealed two related calycin-like domains with structural similarity to the mammalian immune modulatory protein, lipocalin-2. As a secreted bacterial protein required by both partners for optimal fitness, AimA is an exemplar bacterial mutualism factor.
Organizational Affiliation:
Institute of Molecular Biology, University of Oregon, Eugene, United States.