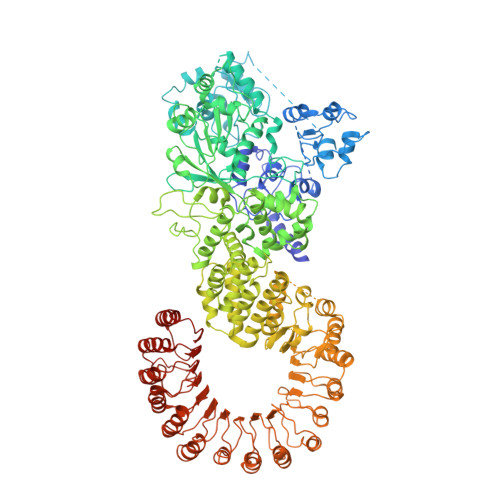
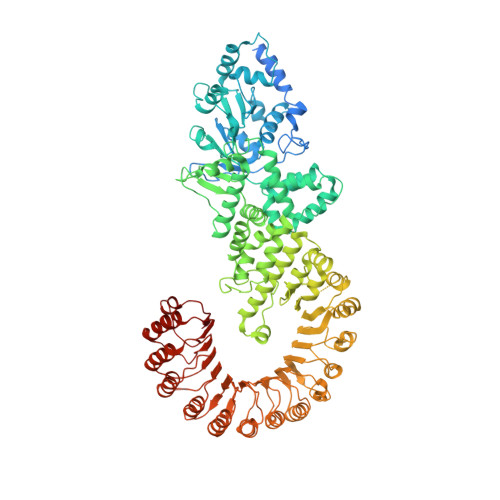
The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion.
Tenthorey, J.L., Haloupek, N., Lopez-Blanco, J.R., Grob, P., Adamson, E., Hartenian, E., Lind, N.A., Bourgeois, N.M., Chacon, P., Nogales, E., Vance, R.E.(2017) Science 358: 888-893
- PubMed: 29146805
- DOI: https://doi.org/10.1126/science.aao1140
- Primary Citation of Related Structures:
6B5B - PubMed Abstract:
Robust innate immune detection of rapidly evolving pathogens is critical for host defense. Nucleotide-binding domain leucine-rich repeat (NLR) proteins function as cytosolic innate immune sensors in plants and animals. However, the structural basis for ligand-induced NLR activation has so far remained unknown. NAIP5 (NLR family, apoptosis inhibitory protein 5) binds the bacterial protein flagellin and assembles with NLRC4 to form a multiprotein complex called an inflammasome. Here we report the cryo-electron microscopy structure of the assembled ~1.4-megadalton flagellin-NAIP5-NLRC4 inflammasome, revealing how a ligand activates an NLR. Six distinct NAIP5 domains contact multiple conserved regions of flagellin, prying NAIP5 into an open and active conformation. We show that innate immune recognition of multiple ligand surfaces is a generalizable strategy that limits pathogen evolution and immune escape.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.