Structural and Functional Studies of a Pyran Synthase Domain from a trans-Acyltransferase Assembly Line.
Wagner, D.T., Zhang, Z., Meoded, R.A., Cepeda, A.J., Piel, J., Keatinge-Clay, A.T.(2018) ACS Chem Biol 13: 975-983
- PubMed: 29481043
- DOI: https://doi.org/10.1021/acschembio.8b00049
- Primary Citation of Related Structures:
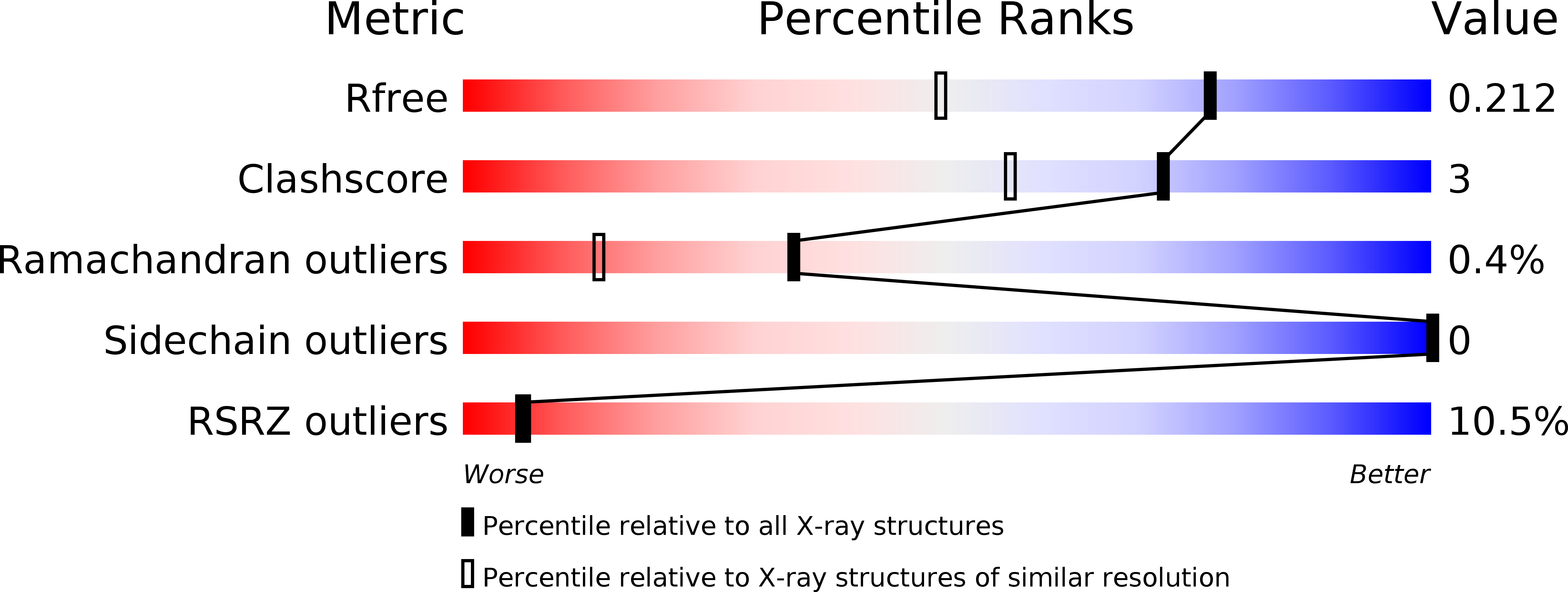
6B2V - PubMed Abstract:
trans-Acyltransferase assembly lines possess enzymatic domains often not observed in their better characterized cis-acyltransferase counterparts. Within this repertoire of largely unexplored biosynthetic machinery is a class of enzymes called the pyran synthases that catalyze the formation of five- and six-membered cyclic ethers from diverse polyketide chains. The 1.55 Å resolution crystal structure of a pyran synthase domain excised from the ninth module of the sorangicin assembly line highlights the similarity of this enzyme to the ubiquitous dehydratase domain and provides insight into the mechanism of ring formation. Functional assays of point mutants reveal the central importance of the active site histidine that is shared with the dehydratases as well as the supporting role of a neighboring semiconserved asparagine.
Organizational Affiliation:
Department of Molecular Biosciences , The University of Texas at Austin , Austin , Texas 78712 , United States.