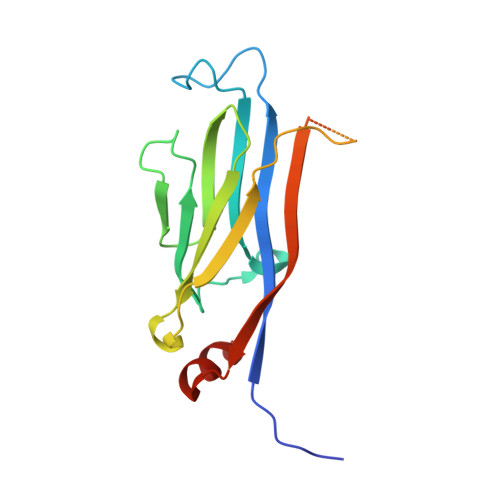
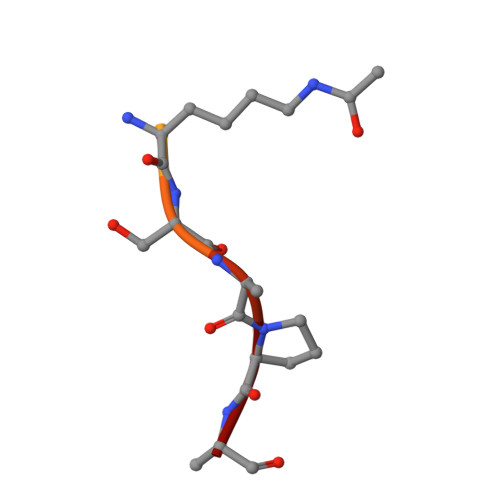
Yaf9 subunit of the NuA4 and SWR1 complexes targets histone H3K27ac through its YEATS domain.
Klein, B.J., Ahmad, S., Vann, K.R., Andrews, F.H., Mayo, Z.A., Bourriquen, G., Bridgers, J.B., Zhang, J., Strahl, B.D., Cote, J., Kutateladze, T.G.(2018) Nucleic Acids Res 46: 421-430
- PubMed: 29145630
- DOI: https://doi.org/10.1093/nar/gkx1151
- Primary Citation of Related Structures:
6AXJ - PubMed Abstract:
Yaf9 is an integral part of the NuA4 acetyltransferase and the SWR1 chromatin remodeling complexes. Here, we show that Yaf9 associates with acetylated histone H3 with high preference for H3K27ac. The crystal structure of the Yaf9 YEATS domain bound to the H3K27ac peptide reveals that the sequence C-terminal to K27ac stabilizes the complex. The side chain of K27ac inserts between two aromatic residues, mutation of which abrogates the interaction in vitro and leads in vivo to phenotypes similar to YAF9 deletion, including loss of SWR1-dependent incorporation of variant histone H2A.Z. Our findings reveal the molecular basis for the recognition of H3K27ac by a YEATS reader and underscore the importance of this interaction in mediating Yaf9 function within the NuA4 and SWR1 complexes.
Organizational Affiliation:
Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.