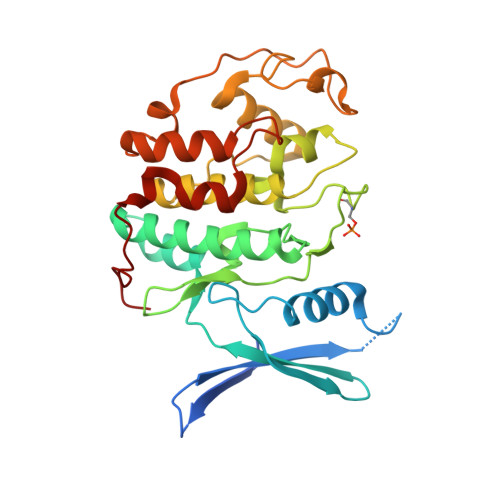
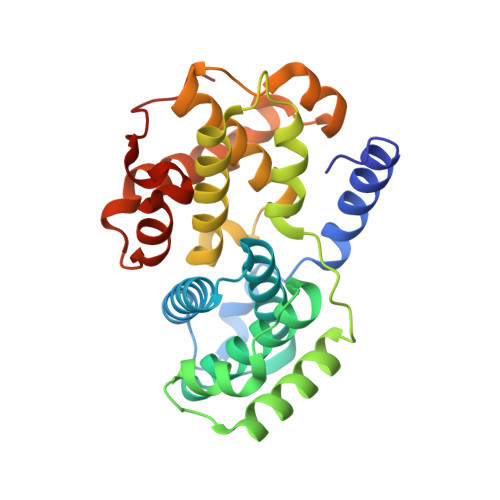
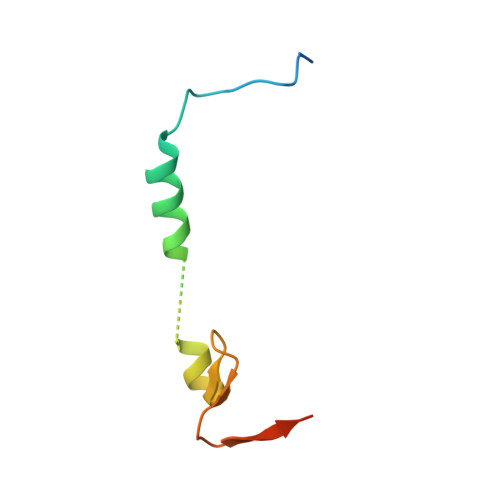
Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation.
Tsytlonok, M., Sanabria, H., Wang, Y., Felekyan, S., Hemmen, K., Phillips, A.H., Yun, M.K., Waddell, M.B., Park, C.G., Vaithiyalingam, S., Iconaru, L., White, S.W., Tompa, P., Seidel, C.A.M., Kriwacki, R.(2019) Nat Commun 10: 1676-1676
- PubMed: 30976006
- DOI: https://doi.org/10.1038/s41467-019-09446-w
- Primary Citation of Related Structures:
6ATH - PubMed Abstract:
p27 Kip1 is an intrinsically disordered protein (IDP) that inhibits cyclin-dependent kinase (Cdk)/cyclin complexes (e.g., Cdk2/cyclin A), causing cell cycle arrest. Cell division progresses when stably Cdk2/cyclin A-bound p27 is phosphorylated on one or two structurally occluded tyrosine residues and a distal threonine residue (T187), triggering degradation of p27. Here, using an integrated biophysical approach, we show that Cdk2/cyclin A-bound p27 samples lowly-populated conformations that provide access to the non-receptor tyrosine kinases, BCR-ABL and Src, which phosphorylate Y88 or Y88 and Y74, respectively, thereby promoting intra-assembly phosphorylation (of p27) on distal T187. Even when tightly bound to Cdk2/cyclin A, intrinsic flexibility enables p27 to integrate and process signaling inputs, and generate outputs including altered Cdk2 activity, p27 stability, and, ultimately, cell cycle progression. Intrinsic dynamics within multi-component assemblies may be a general mechanism of signaling by regulatory IDPs, which can be subverted in human disease.
Organizational Affiliation:
VIB Center for Structural Biology, Vrije Universiteit Brussel, Pleinlaan, 2 1050, Brussels, Belgium.