Structural Insights into Malic Enzyme Variants Favoring an Unnatural Redox Cofactor.
Liu, Y., Guo, X., Liu, W., Wang, J., Zhao, Z.K.(2021) Chembiochem 22: 1765-1768
- PubMed: 33523590
- DOI: https://doi.org/10.1002/cbic.202000800
- Primary Citation of Related Structures:
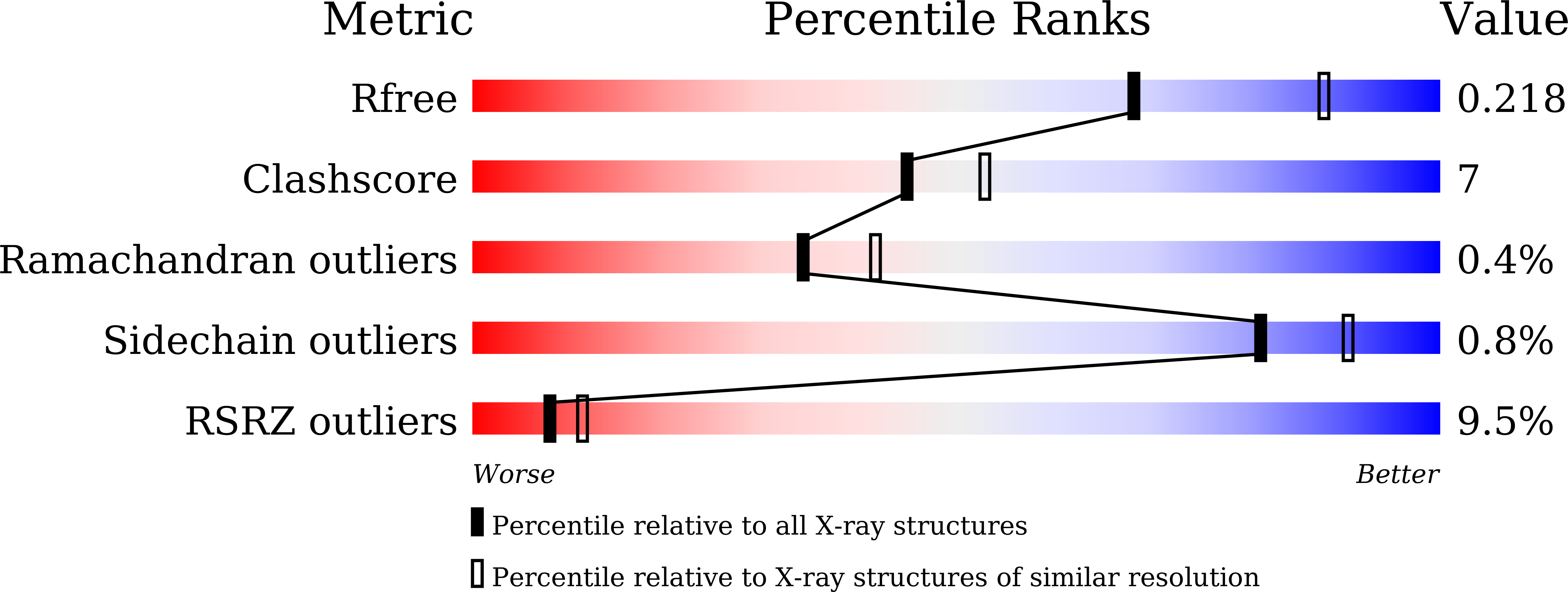
6AGS - PubMed Abstract:
The use of nicotinamide cytosine dinucleotide (NCD), a biocompatible nicotinamide adenosine dinucleotide (NAD) analogue, is of great scientific and biotechnological interest. Several redox enzymes have been devised to favor NCD, and have been successfully applied in creating NCD-dependent redox systems. However, molecular interactions between cofactor and protein have still to be disclosed in order to guide further engineering efforts. Here we report the structural analysis of an NCD-favoring malic enzyme (ME) variant derived from Escherichia coli. The X-ray crystal structure data revealed that the residues located at position 346 and 401 in ME acted as the "gatekeepers" of the adenine moiety binding cavity. When Arg346 was substituted with either acidic or aromatic residues, the corresponding mutants showed substantially reduced NCD preference. Inspired by these observations, we generated Lactobacillus helveticus derived d-lactate dehydrogenase variants at Ile177, the counterpart to Arg346 in ME, and found a similar trend in terms of cofactor preference changes. As many NAD-dependent oxidoreductases share key structural features, our results provide guidance for protein engineering to obtain more NCD-favoring variants.
Organizational Affiliation:
College of Life Sciences, Henan Normal University, 46 East of Construction Road, Xinxiang, 453007, P. R. China.