Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Lee, Y., Yoon, E., Cho, S., Schmahling, S., Muller, J., Song, J.J.(2019) Structure 27: 846
- PubMed: 30827841
- DOI: https://doi.org/10.1016/j.str.2019.01.016
- Primary Citation of Related Structures:
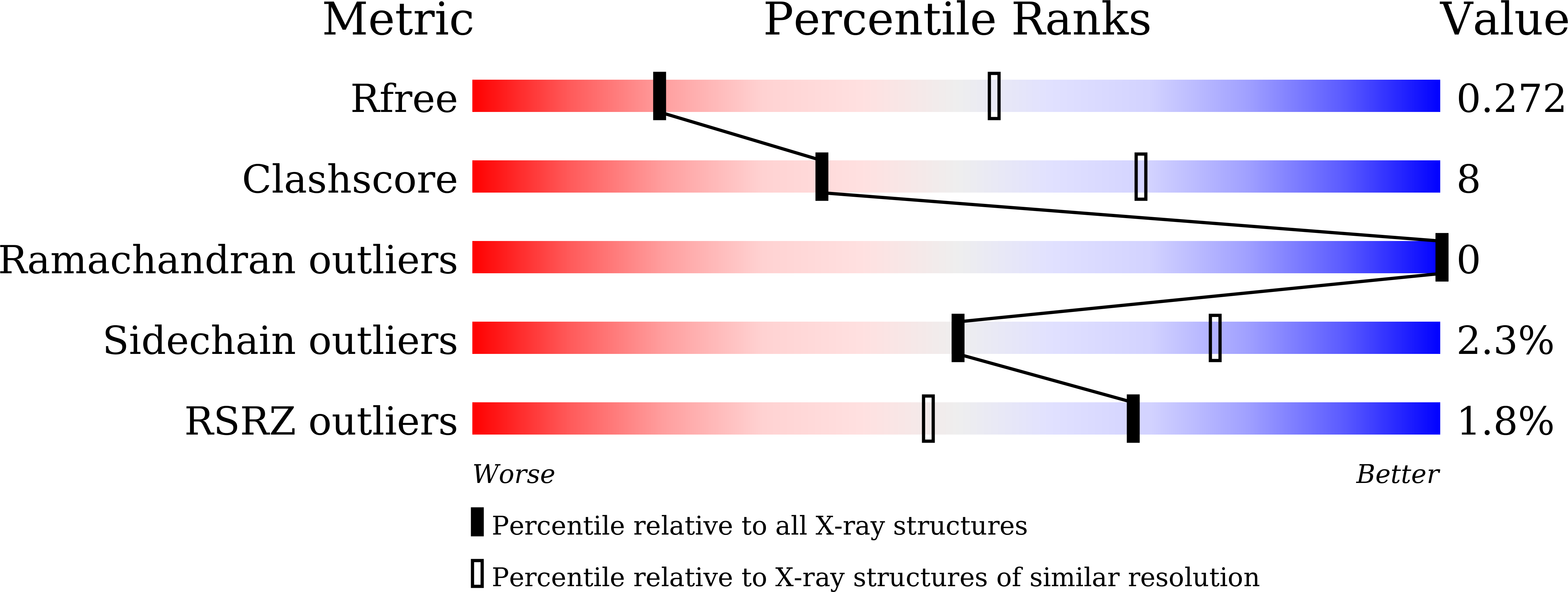
6AGO - PubMed Abstract:
Human ASH1L is the catalytic subunit of the conserved histone methyltransferase (HMTase) complex AMC that dimethylates lysine 36 in histone H3 (H3K36me2) to promote gene transcription in mammals and flies. Unlike AMC, ASH1L alone shows poor catalytic activity, because access to its substrate binding pocket is blocked by an autoinhibitory loop (AI loop) from the postSET domain. We report the crystal structure of the minimal catalytic active AMC complex containing ASH1L and its partner subunit MRG15. The structure reveals how binding of the MRG domain of MRG15 to a conserved FxLP motif in ASH1L results in the displacement of the AI loop to permit substrates to access the catalytic pocket of the ASH1L SET domain. Together, ASH1L activation by MRG15 therefore represents a delicate regulatory mechanism for how a cofactor activates an SET domain HMTase by releasing autoinhibition.
Organizational Affiliation:
Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea; Center for Bioanalysis, Korea Research Institute of Standards and Science, 267 Gajeong-ro, Yuseong-gu, Daejeon 34113, Korea.