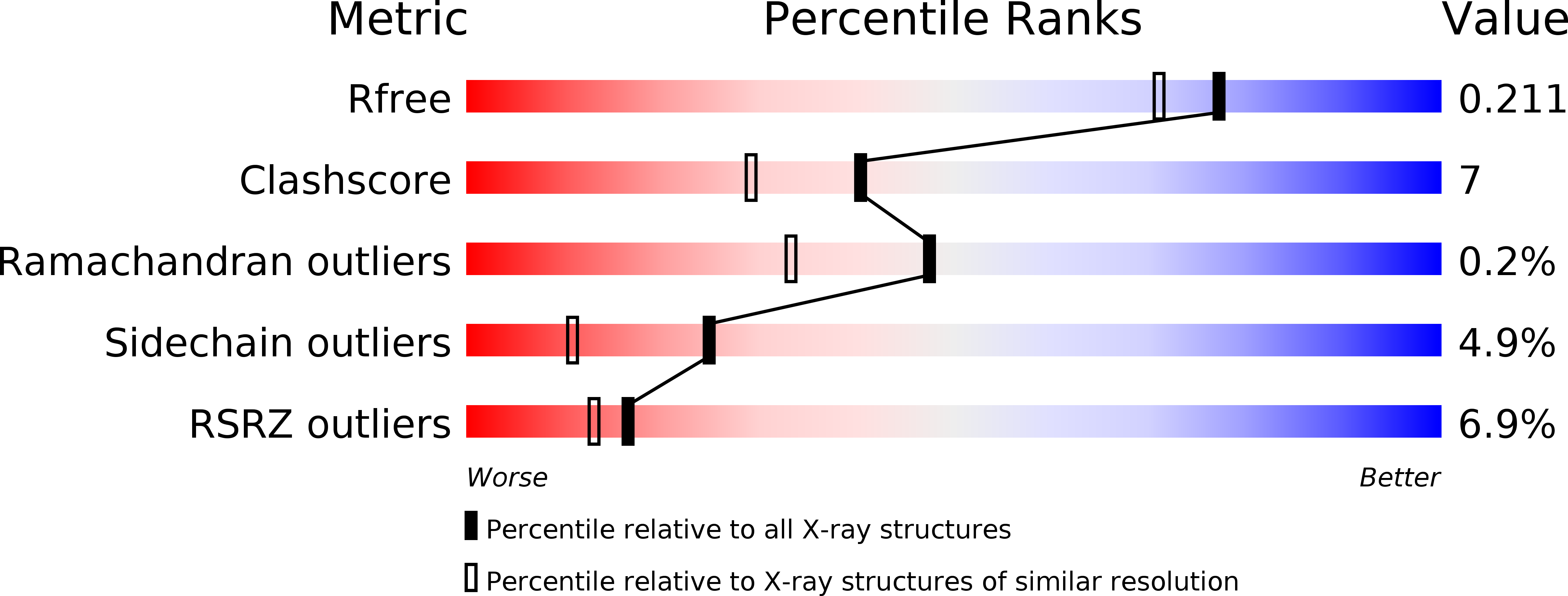
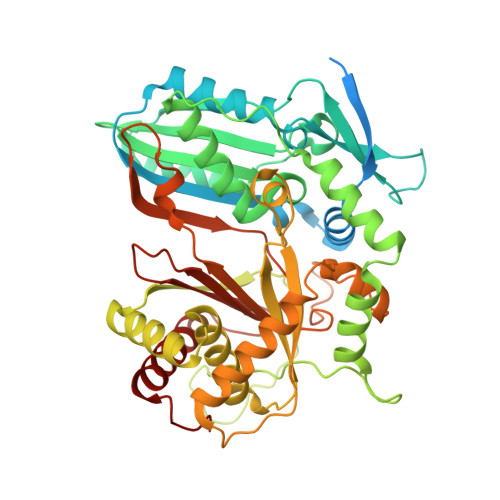
Crystal structure of the condensation domain from lovastatin polyketide synthase.
Wang, L., Yuan, M., Zheng, J.(2019) Synth Syst Biotechnol 4: 10-15
- PubMed: 30533541
- DOI: https://doi.org/10.1016/j.synbio.2018.11.003
- Primary Citation of Related Structures:
6AD3 - PubMed Abstract:
The highly reducing iterative polyketide synthases responsible for lovastatin biosynthesis contains a section homologous to condensation (CON) domain observed in nonribosomal peptide synthetases (NRPSs). In the present study, we expressed the isolated lovastatin CON domain and solved the crystal structure to 1.79 Å resolution. The overall structure shows similarity to canonical condensation domains of NRPSs, containing the N-terminal and C-terminal subdomains that resemble enzymes of chloramphenicol acetyltransferase family, whereas distinct structural features are observed at the active site. The acceptor entry of the substrate channel is blocked by a flexible loop, thereby preventing the loading of substrate for a new round of chain elongation. The mutation of conserved catalytic motif located at the midpoint of substrate channel agrees with the incapability of CON to catalyzed amide-bond formation. The structure helps to understand the function of CON in lovastatin biosynthesis.
Organizational Affiliation:
State Key Laboratory of Microbial Metabolism, and School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.