EhFP10: A FYVE family GEF interacts with myosin IB to regulate cytoskeletal dynamics during endocytosis in Entamoeba histolytica.
Gautam, G., Ali, M.S., Bhattacharya, A., Gourinath, S.(2019) PLoS Pathog 15: e1007573-e1007573
- PubMed: 30779788
- DOI: https://doi.org/10.1371/journal.ppat.1007573
- Primary Citation of Related Structures:
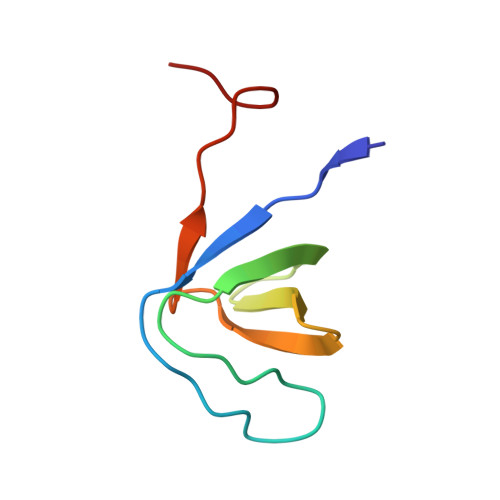
6A9C - PubMed Abstract:
Motility and phagocytosis are key processes that are involved in invasive amoebiasis disease caused by intestinal parasite Entamoeba histolytica. Previous studies have reported unconventional myosins to play significant role in membrane based motility as well as endocytic processes. EhMyosin IB is the only unconventional myosin present in E. histolytica, is thought to be involved in both of these processes. Here, we report an interaction between the SH3 domain of EhMyosin IB and c-terminal domain of EhFP10, a Rho guanine nucleotide exchange factor. EhFP10 was found to be confined to Entamoeba species only, and to contain a c-terminal domain that binds and bundles actin filaments. EhFP10 was observed to localize in the membrane ruffles, phagocytic and macropinocytic cups of E. histolytica trophozoites. It was also found in early pinosomes but not early phagosomes. A crystal structure of the c-terminal SH3 domain of EhMyosin IB (EhMySH3) in complex with an EhFP10 peptide and co-localization studies established the interaction of EhMySH3 with EhFP10. This interaction was shown to lead to inhibition of actin bundling activity and to thereby regulate actin dynamics during endocytosis. We hypothesize that unique domain architecture of EhFP10 might be compensating the absence of Wasp and related proteins in Entamoeba, which are known partners of myosin SH3 domains in other eukaryotes. Our findings also highlights the role of actin bundling during endocytosis.
Organizational Affiliation:
School of Life Sciences, Jawaharlal Nehru University, New Delhi, India.