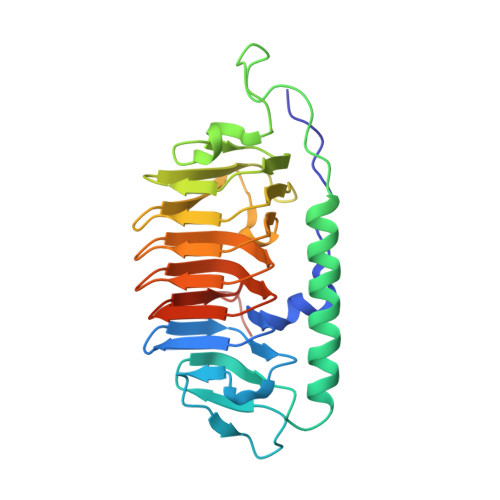
Multiple binding modes of a moderate ice-binding protein from a polar microalga
Kondo, H., Mochizuki, K., Bayer-Giraldi, M.(2018) Phys Chem Chem Phys 20: 25295-25303
- PubMed: 30255887
- DOI: https://doi.org/10.1039/c8cp04727h
- Primary Citation of Related Structures:
6A8K - PubMed Abstract:
Ice-binding proteins (IBPs) produced by cold-tolerant organisms interact with ice and strongly control crystal growth. The molecular basis for the different magnitudes of activity displayed by various IBPs (moderate and hyperactive) has not yet been clarified. Previous studies questioned whether the moderate activity of some IBPs relies on their weaker binding modus to the ice surface, compared to hyperactive IBPs, rather than relying on binding only to selected faces of the ice crystal. We present the structure of one moderate IBP from the sea-ice diatom Fragilariopsis cylindrus (fcIBP) as determined by X-ray crystallography and investigate the protein's binding modes to the growing ice-water interface using molecular dynamics simulations. The structure of fcIBP is the IBP-1 fold, defined by a discontinuous β-solenoid delimitated by three faces (A, B and C-faces) and braced by an α-helix. The fcIBP structure shows capping loops on both N- and C-terminal parts of the solenoid. We show that the protein adsorbs on both the prism and the basal faces of ice crystals, confirming experimental results. The fcIBP binds irreversibly to the prism face using the loop between the B and the C-faces, involving also the B-face in water immobilization despite its irregular structure. The α-helix attaches the protein to the basal face with a partly reversible modus. Our results suggest that fcIBP has a looser attachment to ice and that this weaker binding modus is the basis to explain the moderate activity of fcIBP.
Organizational Affiliation:
Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Sapporo 062-8517, Japan.