The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Jin, H.S., Dhanasingh, I., Sung, J.Y., La, J.W., Lee, Y., Lee, E.M., Kang, Y., Lee, D.Y., Lee, S.H., Lee, D.W.(2020) Microb Biotechnol
- PubMed: 33320434
- DOI: https://doi.org/10.1111/1751-7915.13717
- Primary Citation of Related Structures:
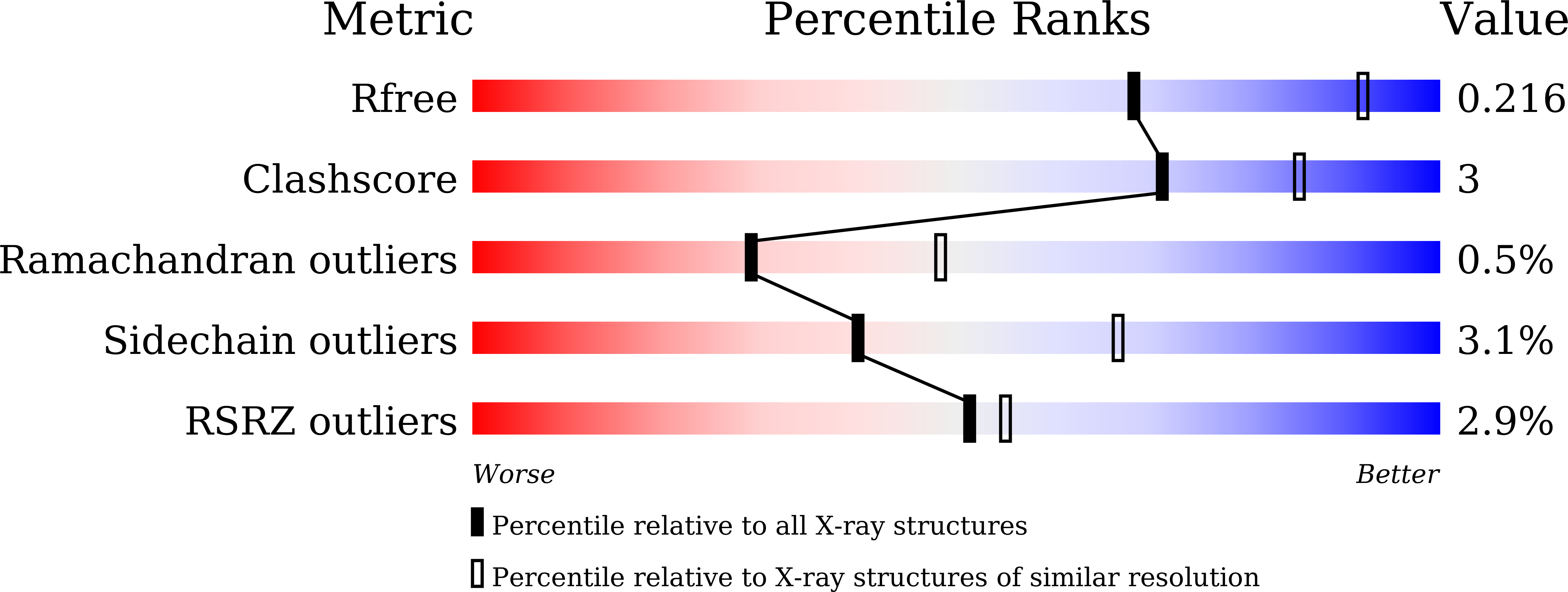
6A6E, 6A6F, 6A6G - PubMed Abstract:
Most extremophilic anaerobes possess a sulfur formation (Suf) system for Fe-S cluster biogenesis. In addition to its essential role in redox chemistry and stress responses of Fe-S cluster proteins, the Suf system may play an important role in keratin degradation by Fervidobacterium islandicum AW-1. Comparative genomics of the order Thermotogales revealed that the feather-degrading F. islandicum AW-1 has a complete Suf-like machinery (SufCBDSU) that is highly expressed in cells grown on native feathers in the absence of elemental sulfur (S 0 ). On the other hand, F. islandicum AW-1 exhibited a significant retardation in the Suf system-mediated keratin degradation in the presence of S 0 . Detailed differential expression analysis of sulfur assimilation machineries unveiled the mechanism by which an efficient sulfur delivery from persulfurated SufS to SufU is achieved during keratinolysis under sulfur starvation. Indeed, addition of SufS-SufU to cell extracts containing keratinolytic proteases accelerated keratin decomposition in vitro under reducing conditions. Remarkably, mass spectrometric analysis of extracellular and intracellular levels of amino acids suggested that redox homeostasis within cells coupled to extracellular cysteine and cystine recycling might be a prerequisite for keratinolysis. Taken together, these results suggest that the Suf-like machinery including the SufS-SufU complex may contribute to sulfur availability for an extracellular reducing environment as well as intracellular redox homeostasis through cysteine released from keratin hydrolysate under starvation conditions.
Organizational Affiliation:
Department of Biotechnology, Yonsei University, Seoul, 03722, South Korea.