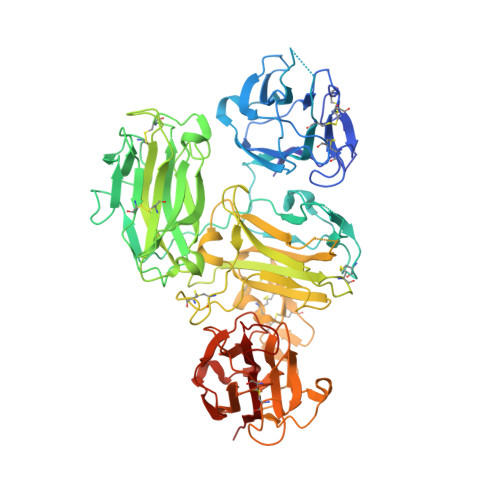
Structural studies of reelin N-terminal region provides insights into a unique structural arrangement and functional multimerization.
Nagae, M., Suzuki, K., Yasui, N., Nogi, T., Kohno, T., Hattori, M., Takagi, J.(2021) J Biochem 169: 555-564
- PubMed: 33377147
- DOI: https://doi.org/10.1093/jb/mvaa144
- Primary Citation of Related Structures:
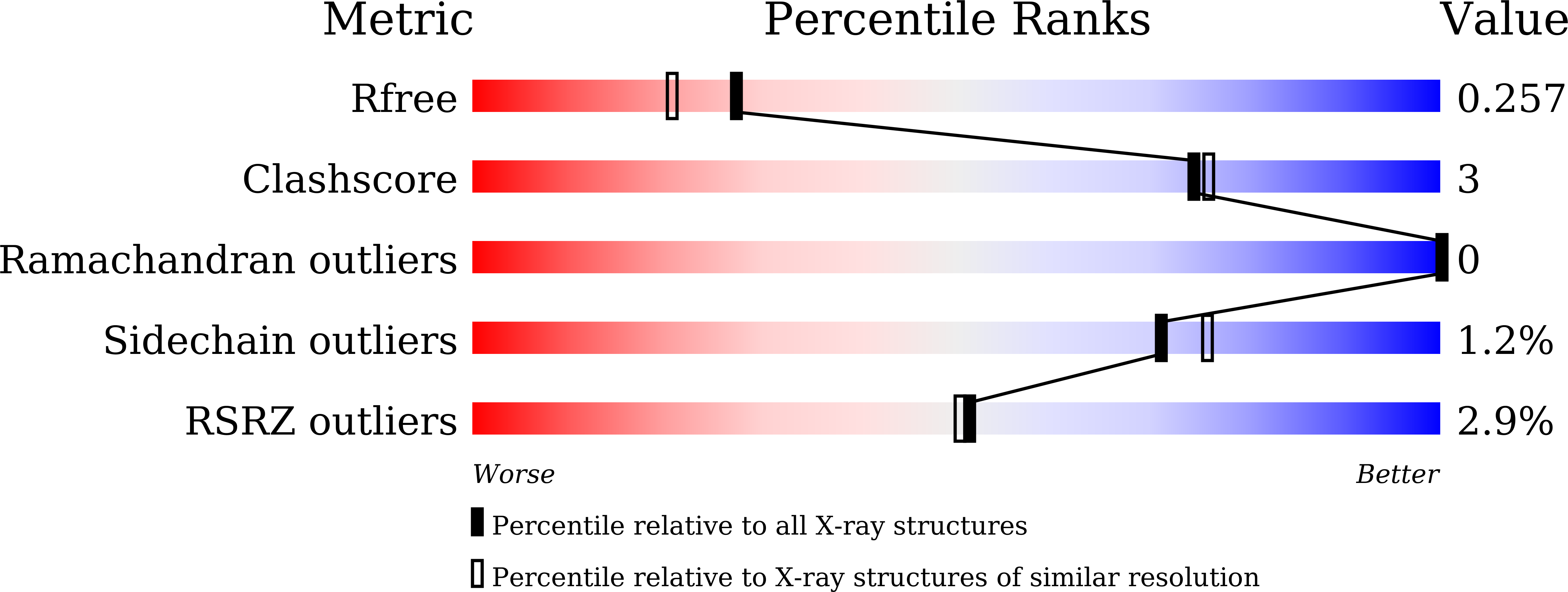
6A48 - PubMed Abstract:
The large, secreted glycoprotein reelin regulates embryonic brain development as well as adult brain functions. Although reelin binds to its receptors via its central part, the N-terminal region directs multimer formation and is critical for efficient signal transduction. In fact, the inhibitory antibody CR-50 interacts with the N-terminal region and prevents higher-order multimerization and signalling. Reelin is a multidomain protein in which the central part is composed of eight characteristic repeats, named reelin repeats, each of which is further divided by insertion of a epidermal growth factor (EGF) module into two subrepeats. In contrast, the N-terminal region shows unique 'irregular' domain architecture since it comprises three consecutive subrepeats without the intervening EGF module. Here, we determined the crystal structure of the murine reelin fragment named RX-R1 including the irregular region and the first reelin repeat at 2.0-Å resolution. The overall structure of RX-R1 has a branched Y-shaped form. Interestingly, two incomplete subrepeats cooperatively form one entire subrepeat structure, though an additional subrepeat is inserted between them. We further reveal that Arg335 of RX-R1 is crucial for binding CR-50. A possible self-association mechanism via the N-terminal region is proposed based on our results.
Organizational Affiliation:
Department of Molecular Immunology, Research Institute for Microbial Diseases.