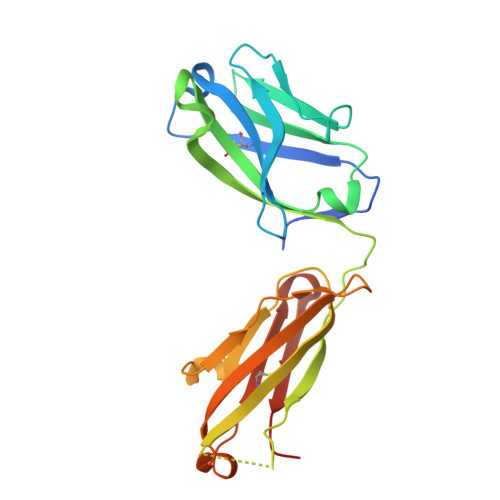
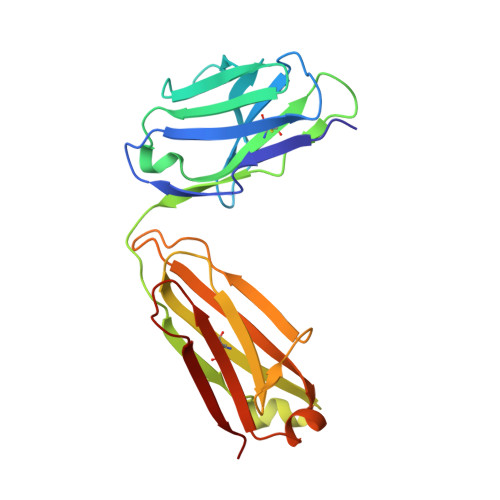
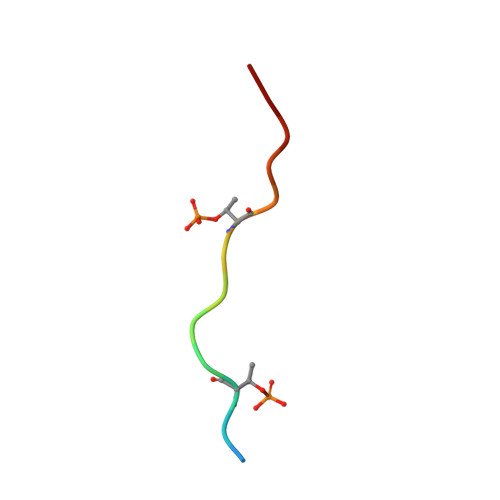
Discovery and Functional Characterization of hPT3, a Humanized Anti-Phospho Tau Selective Monoclonal Antibody.
Van Kolen, K., Malia, T.J., Theunis, C., Nanjunda, R., Teplyakov, A., Ernst, R., Wu, S.J., Luo, J., Borgers, M., Vandermeeren, M., Bottelbergs, A., Wintmolders, C., Lacy, E., Maurin, H., Larsen, P., Willems, R., Van De Casteele, T., Triana-Baltzer, G., Slemmon, R., Galpern, W., Trojanowski, J.Q., Sun, H., Mercken, M.H.(2020) J Alzheimers Dis 77: 1397-1416
- PubMed: 32894244
- DOI: https://doi.org/10.3233/JAD-200544
- Primary Citation of Related Structures:
6XLI - PubMed Abstract:
As a consequence of the discovery of an extracellular component responsible for the progression of tau pathology, tau immunotherapy is being extensively explored in both preclinical and clinical studies as a disease modifying strategy for the treatment of Alzheimer's disease.
Organizational Affiliation:
Neuroscience Department, Janssen Research and Development, Beerse, Belgium.