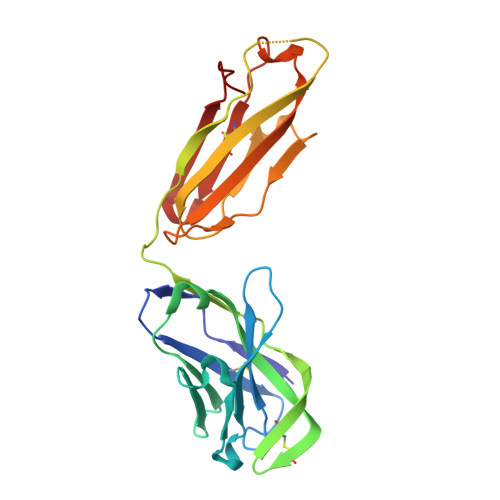
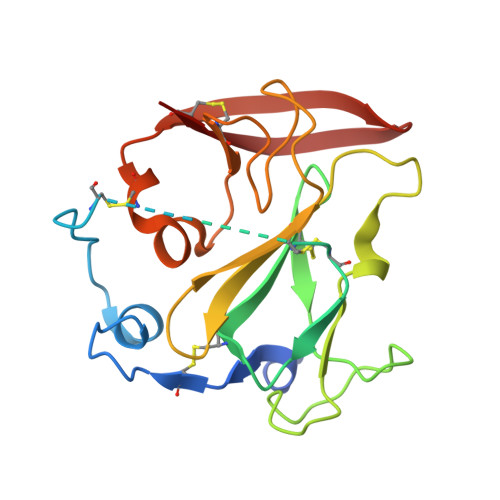
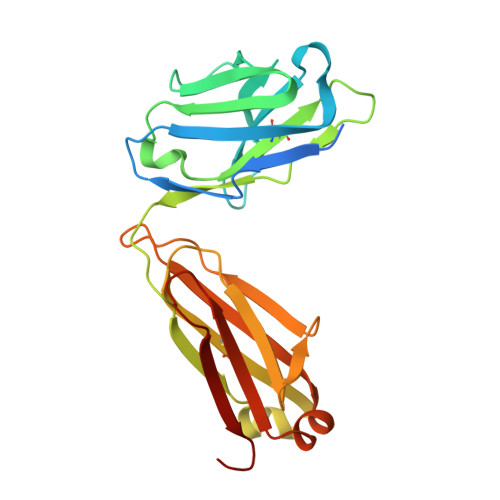
An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Tzarum, N., Giang, E., Kadam, R.U., Chen, F., Nagy, K., Augestad, E.H., Velazquez-Moctezuma, R., Keck, Z.Y., Hua, Y., Stanfield, R.L., Dreux, M., Prentoe, J., Foung, S.K.H., Bukh, J., Wilson, I.A., Law, M.(2020) Sci Adv 6: eabb5642-eabb5642
- PubMed: 32754640
- DOI: https://doi.org/10.1126/sciadv.abb5642
- Primary Citation of Related Structures:
6WO4, 6WOR, 6WOS - PubMed Abstract:
To achieve global elimination of hepatitis C virus (HCV), an effective cross-genotype vaccine is needed. The HCV envelope glycoprotein E2 is the main target for neutralizing antibodies (nAbs), which aid in HCV clearance and protection. E2 is structurally flexible and functions in engaging host receptors. Many nAbs bind to the "neutralizing face" on E2, including several broadly nAbs encoded by the V H 1-69 germline gene family that bind to a similar conformation (A) of this face. Here, a previously unknown conformation (B) of the neutralizing face is revealed in crystal structures of two of four additional E2-V H 1-69 nAb complexes. In this conformation, the E2 front-layer region is displaced upon antibody binding, exposing residues in the back layer for direct antibody interaction. This E2 B structure may represent another conformational state in the viral entry process that is susceptible to antibody neutralization and thus provide a new target for rational vaccine development.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.