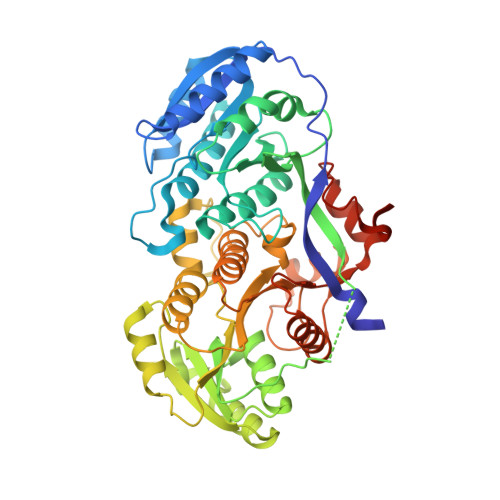
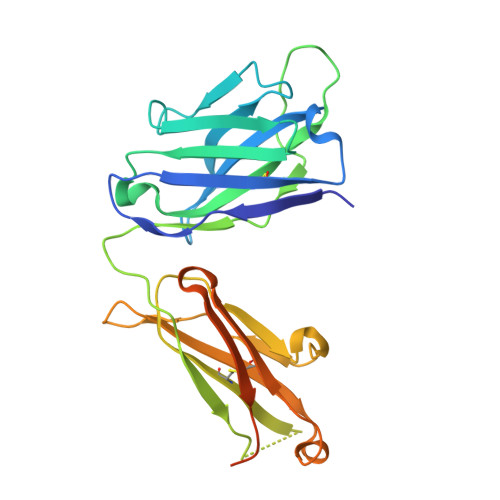
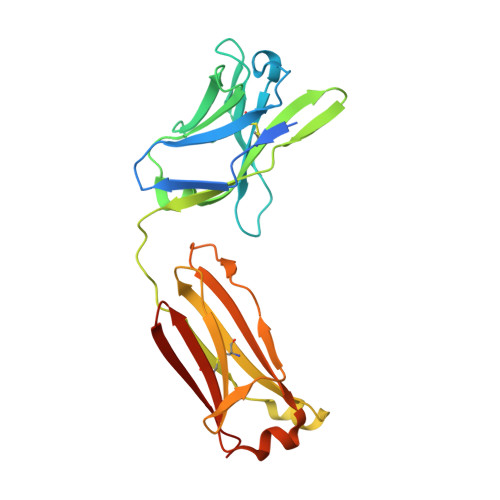
Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
Cogan, D.P., Li, X., Sevillano, N., Mathews, I.I., Matsui, T., Craik, C.S., Khosla, C.(2020) J Am Chem Soc 142: 14933-14939
- PubMed: 32786753
- DOI: https://doi.org/10.1021/jacs.0c05133
- Primary Citation of Related Structures:
6W7S, 6WH9 - PubMed Abstract:
The 6-deoxyerythronolide B synthase (DEBS) is a prototypical assembly line polyketide synthase (PKS) that synthesizes the macrocyclic core of the antibiotic erythromycin. Each of its six multidomain modules presumably sample distinct conformations, as biosynthetic intermediates tethered to their acyl carrier proteins interact with multiple active sites during the courses of their catalytic cycles. The spatiotemporal details underlying these protein dynamics remain elusive. Here, we investigate one aspect of this conformational flexibility using two domain-specific monoclonal antibody fragments (F ab s) isolated from a very large naïve human antibody library. Both F ab s, designated 1D10 and 2G10, were bound specifically and with high affinity to the ketoreductase domain of DEBS module 1 (KR1). Comparative kinetic analysis of stand-alone KR1 as well as a truncated bimodular derivative of DEBS revealed that 1D10 inhibited KR1 activity whereas 2G10 did not. Co-crystal structures of each KR1-F ab complex provided a mechanistic rationale for this difference. A hybrid PKS module harboring KR1 was engineered, whose individual catalytic domains have been crystallographically characterized at high resolution. Size exclusion chromatography coupled to small-angle X-ray scattering (SEC-SAXS) of this hybrid module bound to 1D10 provided further support for the catalytic relevance of the "extended" model of a PKS module. Our findings reinforce the power of monoclonal antibodies as tools to interrogate structure-function relationships of assembly line PKSs.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California San Francisco, San Francisco, California 94158, United States.