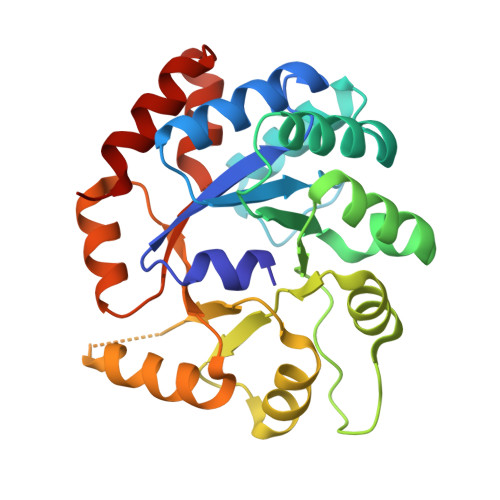
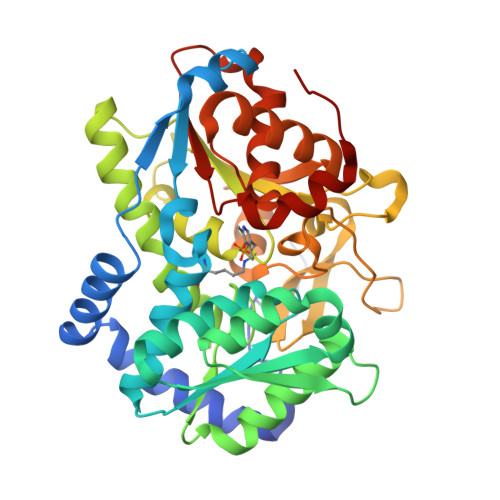
Catalytically impaired TrpA subunit of tryptophan synthase from Chlamydia trachomatis is an allosteric regulator of TrpB.
Michalska, K., Wellington, S., Maltseva, N., Jedrzejczak, R., Selem-Mojica, N., Rosas-Becerra, L.R., Barona-Gomez, F., Hung, D.T., Joachimiak, A.(2021) Protein Sci 30: 1904-1918
- PubMed: 34107106
- DOI: https://doi.org/10.1002/pro.4143
- Primary Citation of Related Structures:
6V82 - PubMed Abstract:
Intracellular growth and pathogenesis of Chlamydia species is controlled by the availability of tryptophan, yet the complete biosynthetic pathway for l-Trp is absent among members of the genus. Some representatives, however, preserve genes encoding tryptophan synthase, TrpAB - a bifunctional enzyme catalyzing the last two steps in l-Trp synthesis. TrpA (subunit α) converts indole-3-glycerol phosphate into indole and glyceraldehyde-3-phosphate (α reaction). The former compound is subsequently used by TrpB (subunit β) to produce l-Trp in the presence of l-Ser and a pyridoxal 5'-phosphate cofactor (β reaction). Previous studies have indicated that in Chlamydia, TrpA has lost its catalytic activity yet remains associated with TrpB to support the β reaction. Here, we provide detailed analysis of the TrpAB from C. trachomatis D/UW-3/CX, confirming that accumulation of mutations in the active site of TrpA renders it enzymatically inactive, despite the conservation of the catalytic residues. We also show that TrpA remains a functional component of the TrpAB complex, increasing the activity of TrpB by four-fold. The side chain of non-conserved βArg267 functions as cation effector, potentially rendering the enzyme less susceptible to the solvent ion composition. The observed structural and functional changes detected herein were placed in a broader evolutionary and genomic context, allowing identification of these mutations in relation to their trp gene contexts in which they occur. Moreover, in agreement with the in vitro data, partial relaxation of purifying selection for TrpA, but not for TrpB, was detected, reinforcing a partial loss of TrpA functions during the course of evolution.
Organizational Affiliation:
Center for Structural Genomics of Infectious Diseases, University of Chicago, Chicago, Illinois, USA.