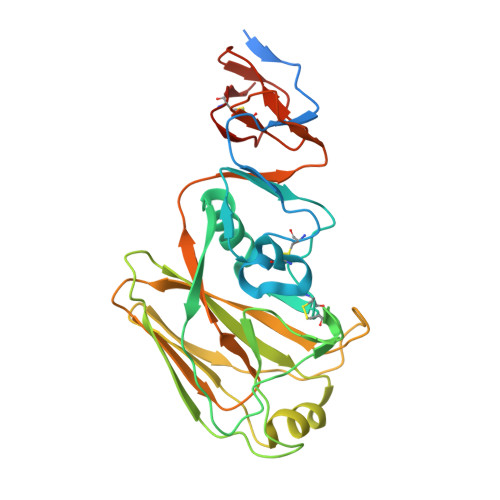
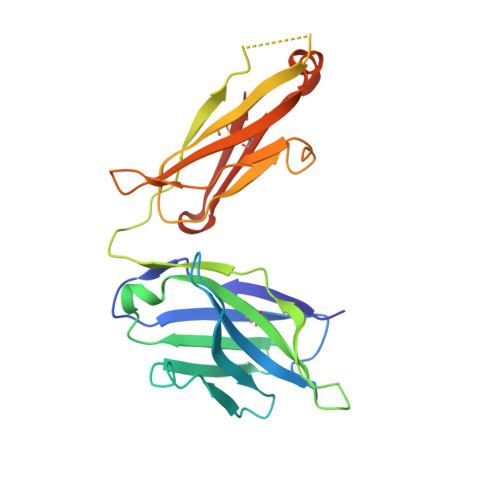
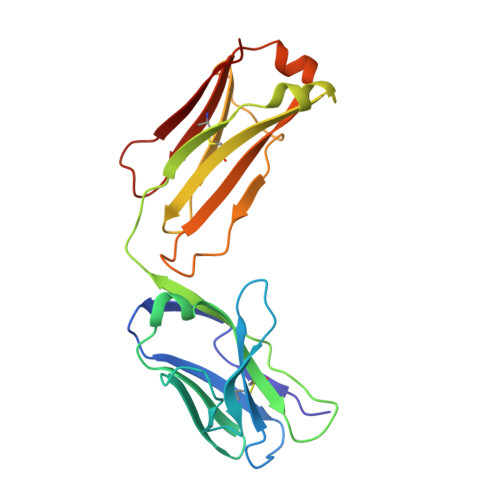
Anti-influenza H7 human antibody targets antigenic site in hemagglutinin head domain interface.
Dong, J., Gilchuk, I., Li, S., Irving, R., Goff, M.T., Turner, H.L., Ward, A.B., Carnahan, R.H., Crowe Jr., J.E.(2020) J Clin Invest 130: 4734-4739
- PubMed: 32749241
- DOI: https://doi.org/10.1172/JCI136032
- Primary Citation of Related Structures:
6UIG - PubMed Abstract:
Although broadly protective, stem-targeted Abs against the influenza A virus hemagglutinin (HA) have been well studied, very limited information is available on Abs that broadly recognize the head domain. We determined the crystal structure of the HA protein of the avian H7N9 influenza virus in complex with a pan-H7, non-neutralizing, protective human Ab. The structure revealed a B cell epitope in the HA head domain trimer interface (TI). This discovery of a second major protective TI epitope supports a model in which uncleaved HA trimers exist on the surface of infected cells in a highly dynamic state that exposes hidden HA head domain features.
Organizational Affiliation:
Vanderbilt Vaccine Center, Vanderbilt Medical Center, Nashville, Tennessee, USA.