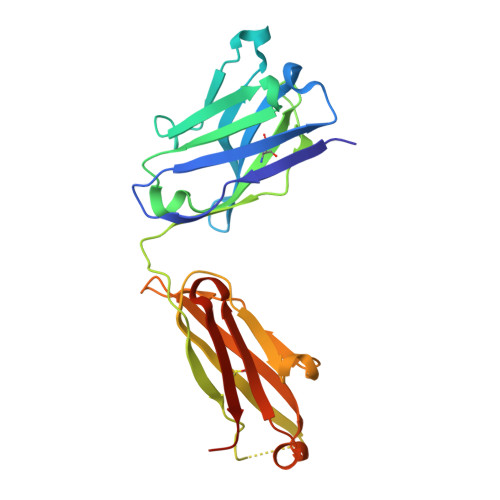
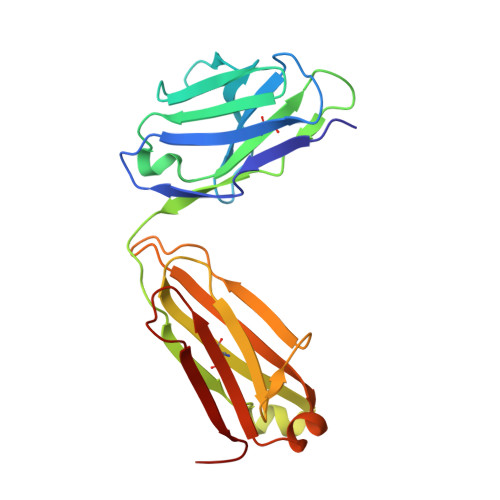
Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
Lerch, T.F., Sharpe, P., Mayclin, S.J., Edwards, T.E., Polleck, S., Rouse, J.C., Zou, Q., Conlon, H.D.(2020) BioDrugs 34: 77-87
- PubMed: 31650490
- DOI: https://doi.org/10.1007/s40259-019-00390-1
- Primary Citation of Related Structures:
6UGS, 6UGT, 6UGU, 6UGV - PubMed Abstract:
Higher-order structure (HOS) assessment is an important component of biosimilarity evaluations. While established spectroscopic methods are routinely used to characterize structure and evaluate similarity, the addition of X-ray crystallographic analysis to these biophysical methods enables orthogonal elucidation of HOS at higher resolution.
Organizational Affiliation:
Analytical Research and Development, Biotherapeutics Pharmaceutical Sciences, Pfizer Inc., Chesterfield, MO, 63017, USA. thomas.lerch@pfizer.com.