Design of Organo-Peptides As Bipartite PCSK9 Antagonists.
Burdick, D.J., Skelton, N.J., Ultsch, M., Beresini, M.H., Eigenbrot, C., Li, W., Zhang, Y., Nguyen, H., Kong-Beltran, M., Quinn, J.G., Kirchhofer, D.(2020) ACS Chem Biol 15: 425-436
- PubMed: 31962046
- DOI: https://doi.org/10.1021/acschembio.9b00899
- Primary Citation of Related Structures:
6U2F, 6U3I - PubMed Abstract:
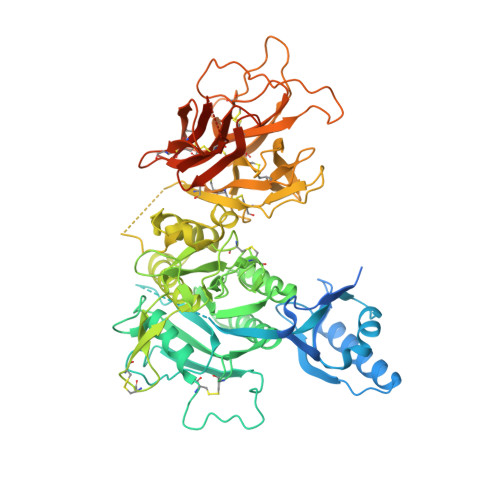
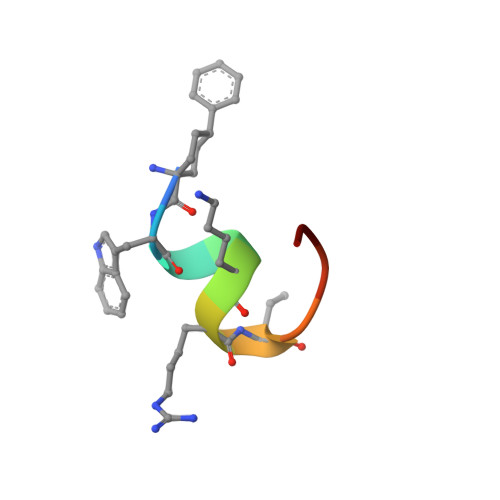
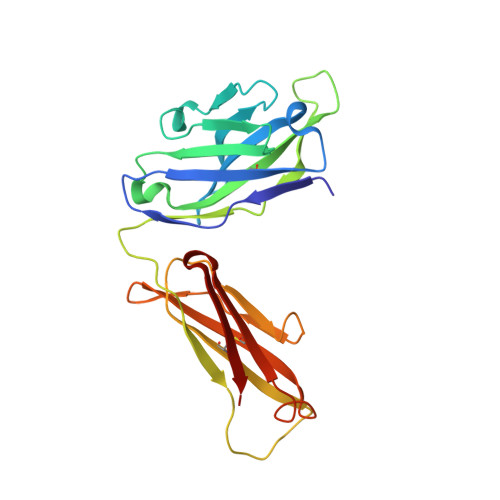
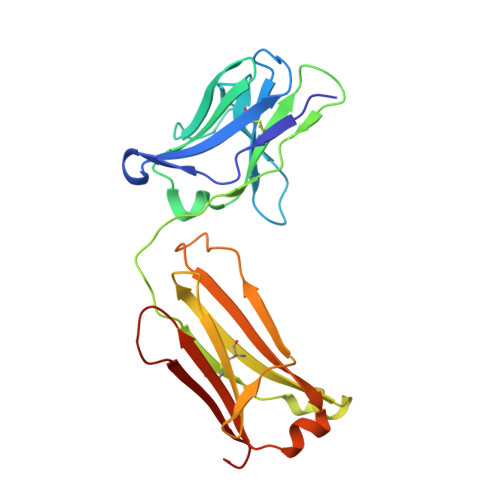
Proprotein convertase subtilisin/kexin 9 (PCSK9) has become an important therapeutic target for lipid lowering, since it regulates low-density lipoprotein cholesterol (LDL-c) levels by binding to liver LDL receptors (LDLR) and effecting their intracellular degradation. However, the development of small molecule inhibitors is hampered by the lack of attractive PCSK9 target sites. We recently discovered helical peptides that are able to bind to a cryptic groove site on PCSK9, which is situated in proximity to the main LDLR binding site. Here, we designed potent bipartite PCSK9 inhibitors by appending organic moieties to a helical groove-binding peptide to reach a hydrophobic pocket in the proximal LDLR binding region. The ultimately designed 1-amino-4-phenylcyclohexane-1-carbonyl extension improved the peptide affinity by >100-fold, yielding organo-peptide antagonists that potently inhibited PCSK9 binding to LDLR and preserved cellular LDLR. These new bipartite antagonists have reduced mass and improved potency compared to the first-generation peptide antagonists, further validating the PCSK9 groove as a viable therapeutic target site.