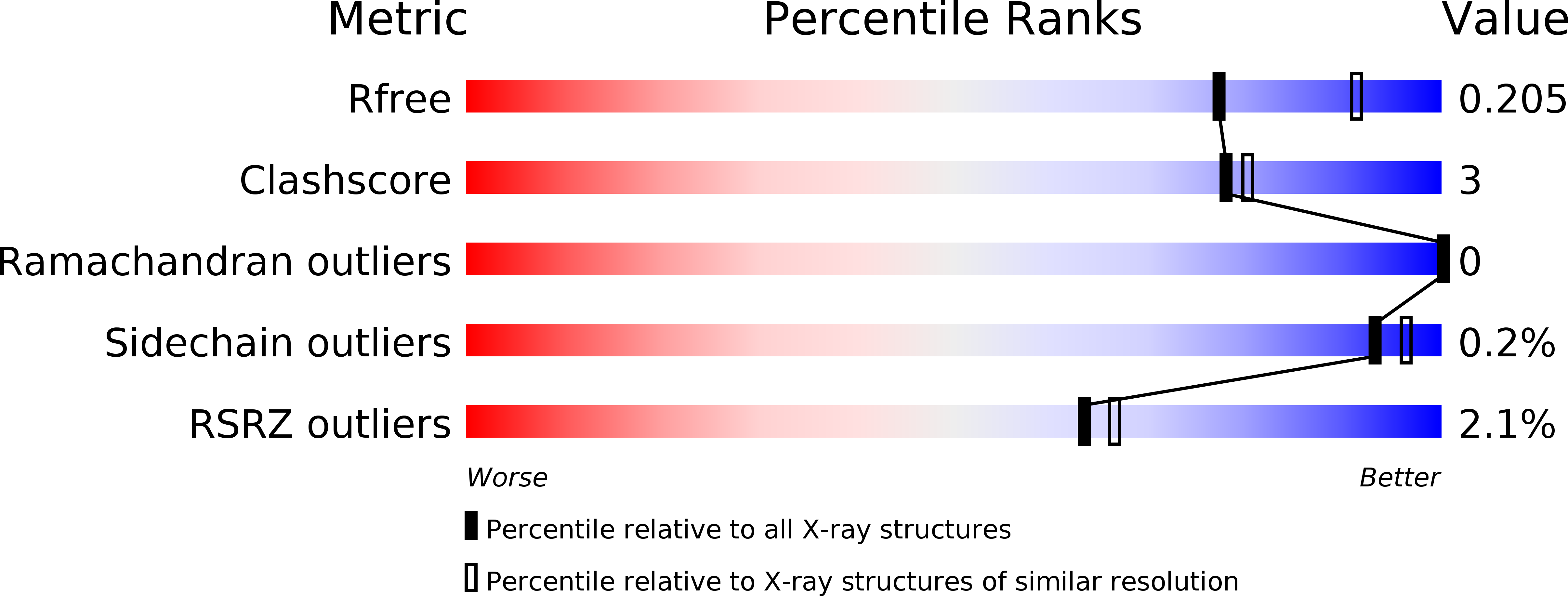
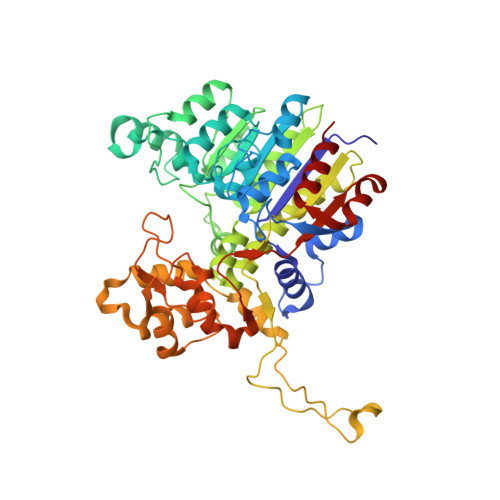
X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine.
Asgeirsson, B., Markusson, S., Hlynsdottir, S.S., Helland, R., Hjorleifsson, J.G.(2020) Biochem Biophys Rep 24: 100830-100830
- PubMed: 33102813
- DOI: https://doi.org/10.1016/j.bbrep.2020.100830
- Primary Citation of Related Structures:
6T26 - PubMed Abstract:
Para -nitrophenyl phosphate, the common substrate for alkaline phosphatase (AP), is available as a cyclohexylamine salt. Here, we report that cyclohexylamine is a non-competitive inhibitor of APs.
Organizational Affiliation:
Department of Biochemistry, Science Institute, University of Iceland, Dunhagi 3, 107 Reykjavik, Iceland.