An Emerin LEM-Domain Mutation Impairs Cell Response to Mechanical Stress.
Essawy, N., Samson, C., Petitalot, A., Moog, S., Bigot, A., Herrada, I., Marcelot, A., Arteni, A.A., Coirault, C., Zinn-Justin, S.(2019) Cells 8
- PubMed: 31185657
- DOI: https://doi.org/10.3390/cells8060570
- Primary Citation of Related Structures:
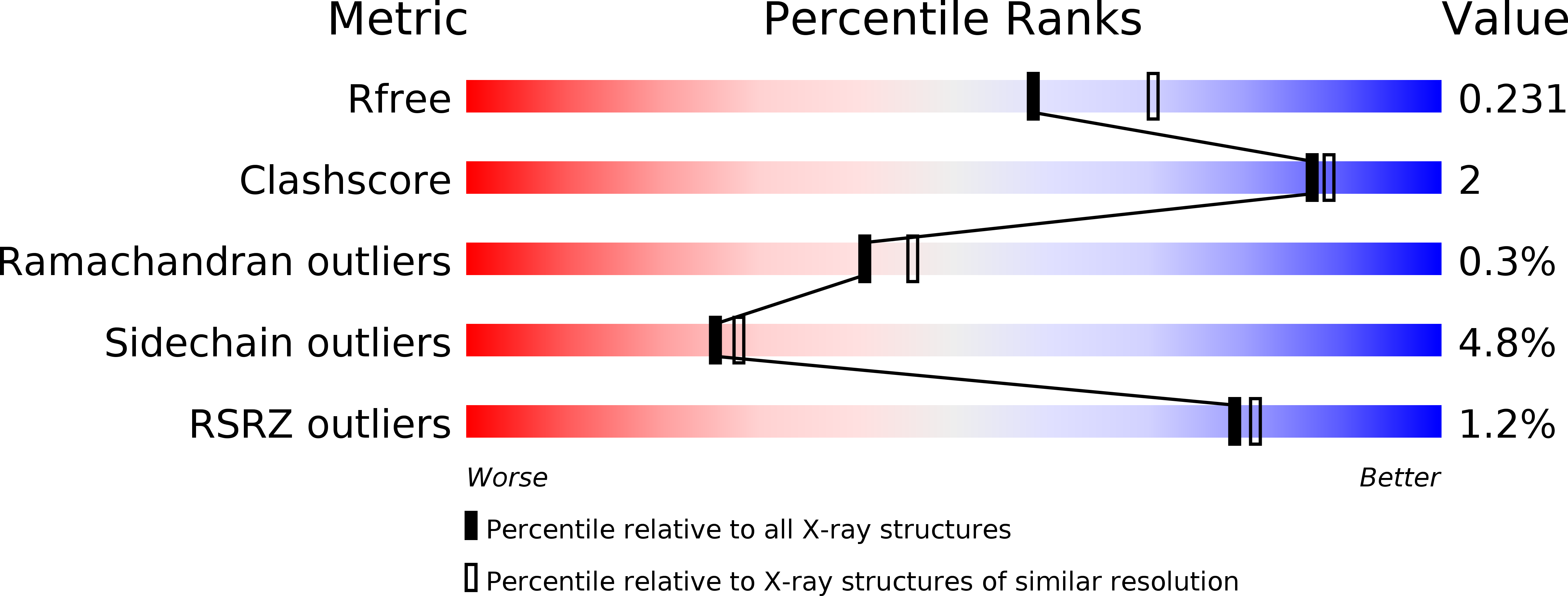
6RPR - PubMed Abstract:
Emerin is a nuclear envelope protein that contributes to genome organization and cell mechanics. Through its N-terminal LAP2-emerin-MAN1 (LEM)-domain, emerin interacts with the DNA-binding protein barrier-to-autointegration (BAF). Emerin also binds to members of the linker of the nucleoskeleton and cytoskeleton (LINC) complex. Mutations in the gene encoding emerin are responsible for the majority of cases of X-linked Emery-Dreifuss muscular dystrophy (X-EDMD). Most of these mutations lead to an absence of emerin. A few missense and short deletion mutations in the disordered region of emerin are also associated with X-EDMD. More recently, missense and short deletion mutations P22L, ∆K37 and T43I were discovered in emerin LEM-domain, associated with isolated atrial cardiac defects (ACD). Here we reveal which defects, at both the molecular and cellular levels, are elicited by these LEM-domain mutations. Whereas K37 mutation impaired the correct folding of the LEM-domain, P22L and T43I had no impact on the 3D structure of emerin. Surprisingly, all three mutants bound to BAF, albeit with a weaker affinity in the case of K37. In human myofibroblasts derived from a patient's fibroblasts, emerin ∆K37 was correctly localized at the inner nuclear membrane, but was present at a significantly lower level, indicating that this mutant is abnormally degraded. Moreover, SUN2 was reduced, and these cells were defective in producing actin stress fibers when grown on a stiff substrate and after cyclic stretches. Altogether, our data suggest that the main effect of mutation K37 is to perturb emerin function within the LINC complex in response to mechanical stress.
Organizational Affiliation:
Sorbonne Université, INSERM UMR_S974, Center for Research in Myology, 75013 Paris, France. nada.elahmady@gmail.com.