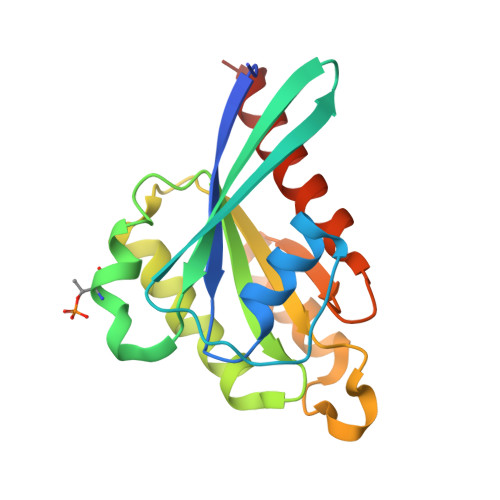
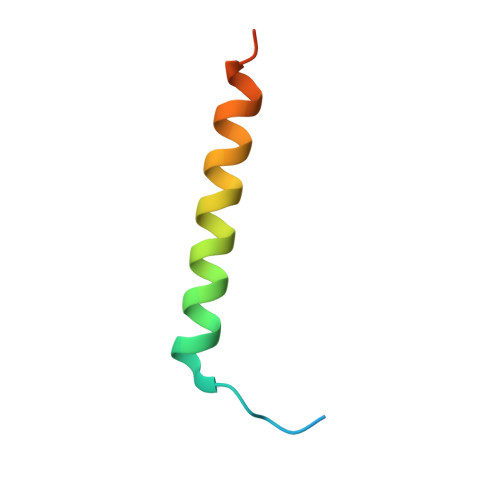
Structural Basis for Rab8a Recruitment of RILPL2 via LRRK2 Phosphorylation of Switch 2.
Waschbusch, D., Purlyte, E., Pal, P., McGrath, E., Alessi, D.R., Khan, A.R.(2020) Structure 28: 406-417.e6
- PubMed: 32017888
- DOI: https://doi.org/10.1016/j.str.2020.01.005
- Primary Citation of Related Structures:
6RIR - PubMed Abstract:
Rab8a is associated with the dynamic regulation of membrane protrusions in polarized cells. Rab8a is one of several Rab GTPases that are substrates of leucine-rich repeat kinase 2 (LRRK2), a serine/threonine kinase that is linked to Parkinson's disease. Rab8a is phosphorylated at T72 (pT72) in its switch 2 helix and recruits the phospho-specific effector RILPL2, which subsequently regulates ciliogenesis. Here, we report the crystal structure of phospho-Rab8a (pRab8a) in complex with the RH2 (RILP homology) domain of RILPL2. The complex is a heterotetramer with RILPL2 forming a central α-helical dimer that bridges two pRab8a molecules. The N termini of the α helices cross over, forming an X-shaped cap (X-cap) that orients Arg residues from RILPL2 toward pT72. X-cap residues critical for pRab8a binding are conserved in JIP3 and JIP4, which also interact with LRRK2-phosphorylated Rab10. We propose a general mode of recognition for phosphorylated Rab GTPases by this family of phospho-specific effectors.
Organizational Affiliation:
School of Biochemistry and Immunology, Trinity College, Dublin 2, Ireland.