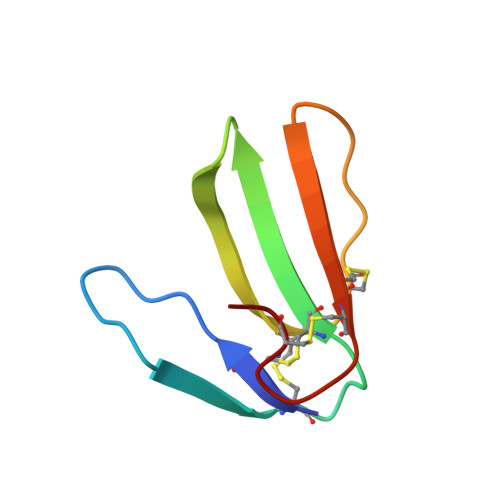
MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Ciolek, J., Zoukimian, C., Dhot, J., Burban, M., Triquigneaux, M., Lauzier, B., Guimbert, C., Boturyn, D., Ferron, M., Ciccone, L., Tepshi, L., Stura, E., Legrand, P., Robin, P., Mourier, G., Schaack, B., Fellah, I., Blanchet, G., Gauthier-Erfanian, C., Beroud, R., Servent, D., De Waard, M., Gilles, N.(2022) Biomed Pharmacother 150: 113094