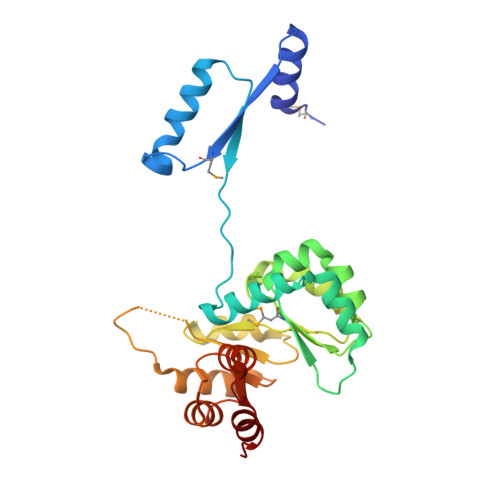
3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Michalska, K., Kowiel, M., Bigelow, L., Endres, M., Gilski, M., Jaskolski, M., Joachimiak, A.(2020) Acta Crystallogr D Struct Biol 76: 166-175
- PubMed: 32038047
- DOI: https://doi.org/10.1107/S2059798320000212
- Primary Citation of Related Structures:
6QKY - PubMed Abstract:
Tryptophan synthase catalyzes the last two steps of tryptophan biosynthesis in plants, fungi and bacteria. It consists of two protein chains, designated α and β, encoded by trpA and trpB genes, that function as an αββα complex. Structural and functional features of tryptophan synthase have been extensively studied, explaining the roles of individual residues in the two active sites in catalysis and allosteric regulation. TrpA serves as a model for protein-folding studies. In 1969, Jackson and Yanofsky observed that the typically monomeric TrpA forms a small population of dimers. Dimerization was postulated to take place through an exchange of structural elements of the monomeric chains, a phenomenon later termed 3D domain swapping. The structural details of the TrpA dimer have remained unknown. Here, the crystal structure of the Streptococcus pneumoniae TrpA homodimer is reported, demonstrating 3D domain swapping in a TIM-barrel fold for the first time. The N-terminal domain comprising the H0-S1-H1-S2 elements is exchanged, while the hinge region corresponds to loop L2 linking strand S2 to helix H2'. The structural elements S2 and L2 carry the catalytic residues Glu52 and Asp63. As the S2 element is part of the swapped domain, the architecture of the catalytic apparatus in the dimer is recreated from two protein chains. The homodimer interface overlaps with the α-β interface of the tryptophan synthase αββα heterotetramer, suggesting that the 3D domain-swapped dimer cannot form a complex with the β subunit. In the crystal, the dimers assemble into a decamer comprising two pentameric rings.
Organizational Affiliation:
Midwest Center for Structural Genomics, X-ray Science Division, Argonne National Laboratory, Argonne, IL 60439, USA.