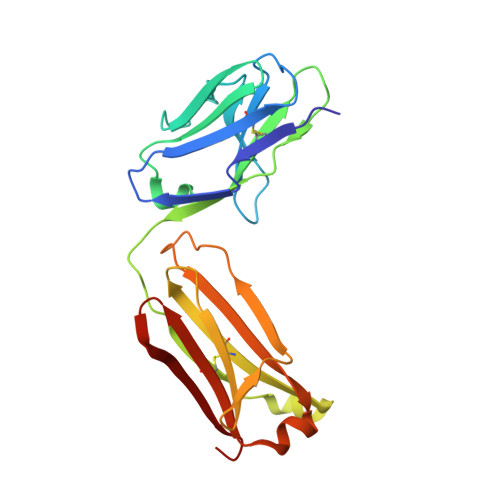
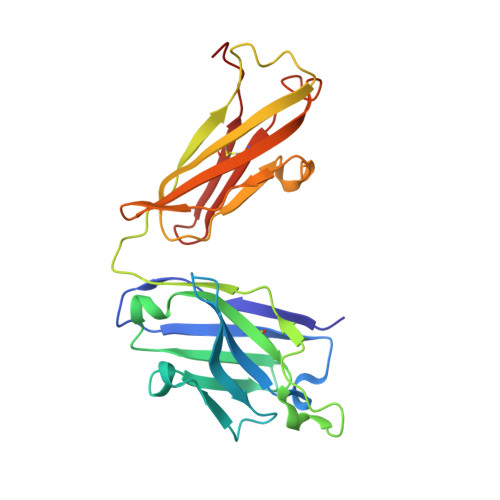
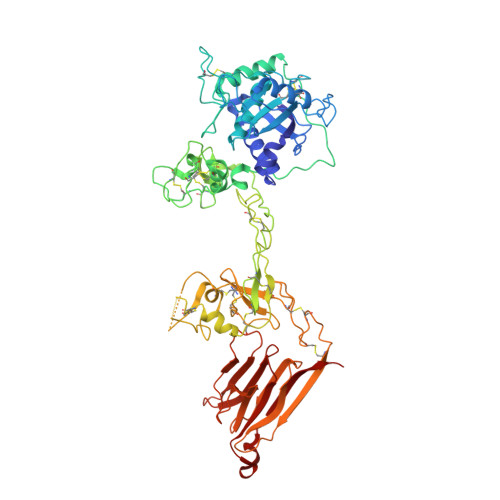
Crystal structure and substrate-induced activation of ADAMTS13.
Petri, A., Kim, H.J., Xu, Y., de Groot, R., Li, C., Vandenbulcke, A., Vanhoorelbeke, K., Emsley, J., Crawley, J.T.B.(2019) Nat Commun 10: 3781-3781
- PubMed: 31439947
- DOI: https://doi.org/10.1038/s41467-019-11474-5
- Primary Citation of Related Structures:
6QIG - PubMed Abstract:
Platelet recruitment to sites of blood vessel damage is highly dependent upon von Willebrand factor (VWF). VWF platelet-tethering function is proteolytically regulated by the metalloprotease ADAMTS13. Proteolysis depends upon shear-induced conformational changes in VWF that reveal the A2 domain cleavage site. Multiple ADAMTS13 exosite interactions are involved in recognition of the unfolded A2 domain. Here we report through kinetic analyses that, in binding VWF, the ADAMTS13 cysteine-rich and spacer domain exosites bring enzyme and substrate into proximity. Thereafter, binding of the ADAMTS13 disintegrin-like domain exosite to VWF allosterically activates the adjacent metalloprotease domain to facilitate proteolysis. The crystal structure of the ADAMTS13 metalloprotease to spacer domains reveals that the metalloprotease domain exhibits a latent conformation in which the active-site cleft is occluded supporting the requirement for an allosteric change to enable accommodation of the substrate. Our data demonstrate that VWF functions as both the activating cofactor and substrate for ADAMTS13.
Organizational Affiliation:
Centre for Haematology, Imperial College London, London, UK.