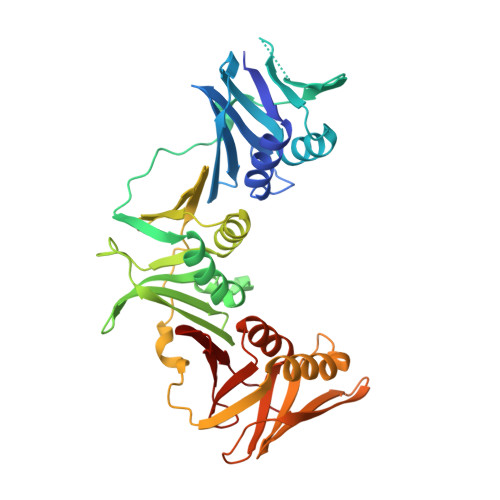
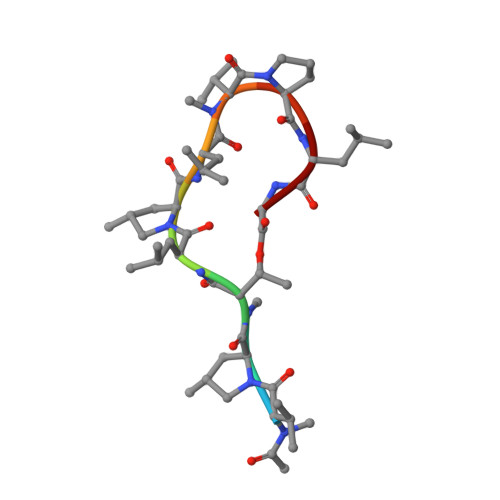
Crystal structure of a DnaN sliding clamp (DNA polymerase III subunit beta) from Bartonella birtlesii bound to griselimycin
Edwards, T.E., Abendroth, J., Horanyi, P.S., Lorimer, D.D., Seattle Structural Genomics Center for Infectious Disease (SSGCID)To be published.