Structure of RNA polymerase binding protein and transcriptional regulator Dks from Chlamydia trachomatis L2 (LGV434)
Abendroth, J., Buchko, G.W., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DnaK Suppressor | 132 | Chlamydia trachomatis | Mutation(s): 0 | 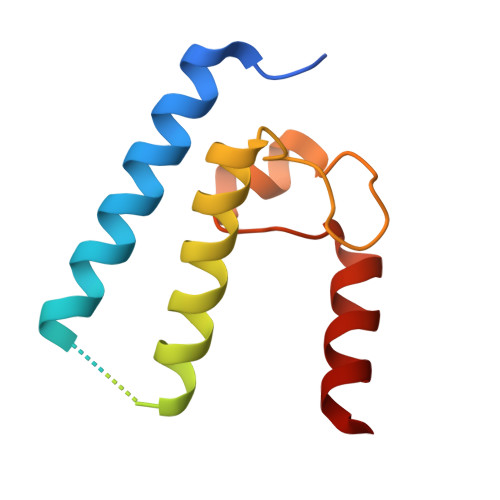 | |
UniProt | |||||
Find proteins for O84412 (Chlamydia trachomatis (strain D/UW-3/Cx)) Explore O84412 Go to UniProtKB: O84412 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O84412 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 83.42 | α = 90 |
| b = 83.42 | β = 90 |
| c = 99.03 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| MR-Rosetta | phasing |
| Coot | model building |