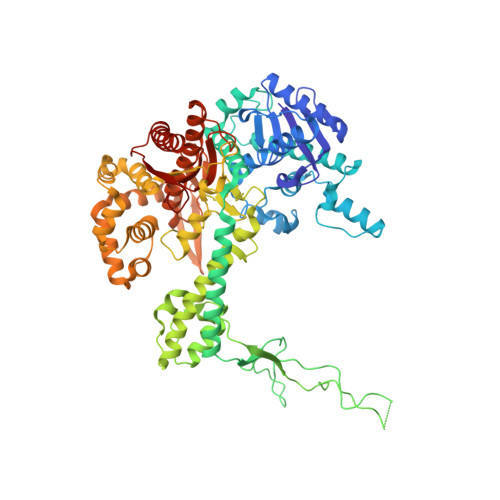
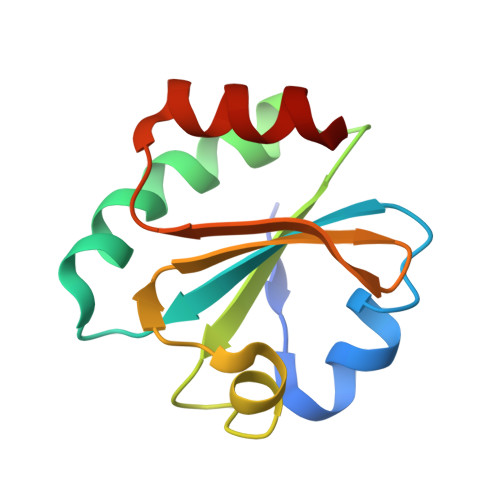
Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Foster, B.M., Rosenberg, D., Salvo, H., Stephens, K.L., Bintz, B.J., Hammel, M., Ellenberger, T., Gainey, M.D., Wallen, J.R.(2019) Biochemistry 58: 4466-4479
- PubMed: 31659895
- DOI: https://doi.org/10.1021/acs.biochem.9b00525
- Primary Citation of Related Structures:
6P7E - PubMed Abstract:
Recent structural studies of the bacteriophage T7 DNA replication system have shed light on how multiple proteins assemble to copy two antiparallel DNA strands. In T7, acidic C-terminal tails of both the primase-helicase and single-stranded DNA binding protein bind to two basic patches on the DNA polymerase to aid in replisome assembly, processivity, and coordinated DNA synthesis. Although these electrostatic interactions are essential for DNA replication, the molecular details for how these tails bind the polymerase are unknown. We have determined an X-ray crystal structure of the T7 DNA polymerase bound to both a primer/template DNA and a peptide that mimics the C-terminal tail of the primase-helicase. The structure reveals that the essential C-terminal phenylalanine of the tail binds to a hydrophobic pocket that is surrounded by positive charge on the surface of the polymerase. We show that alterations of polymerase residues that engage the tail lead to defects in viral replication. In the structure, we also observe dTTP bound in the exonuclease active site and stacked against tryptophan 160. Using both primer/extension assays and high-throughput sequencing, we show how mutations in the exonuclease active site lead to defects in mismatch repair and an increase in the level of mutagenesis of the T7 genome. Finally, using small-angle X-ray scattering, we provide the first solution structures of a complex between the single-stranded DNA binding protein and the DNA polymerase and show how a single-stranded DNA binding protein dimer engages both one and two copies of DNA polymerase.
Organizational Affiliation:
Department of Chemistry & Physics , Western Carolina University , Cullowhee , North Carolina 28723 , United States.