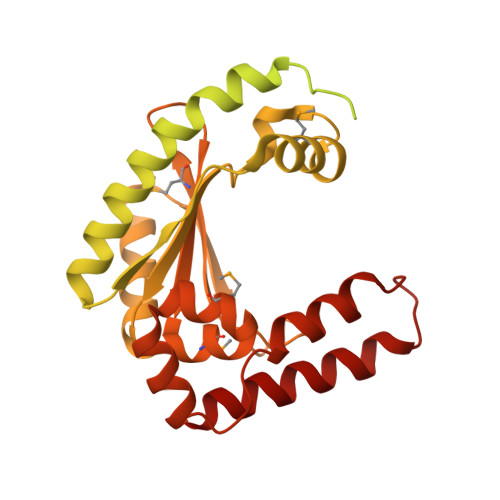
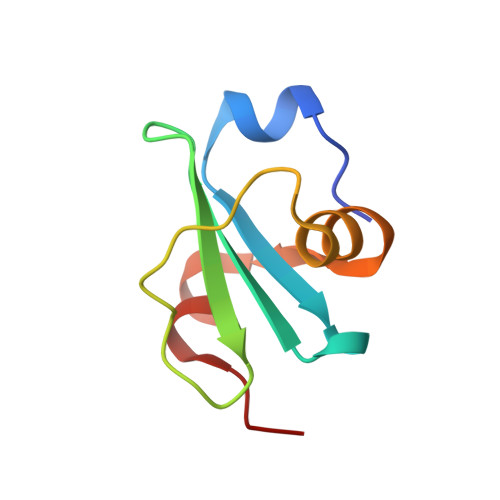
An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp.
Ahmad, S., Wang, B., Walker, M.D., Tran, H.R., Stogios, P.J., Savchenko, A., Grant, R.A., McArthur, A.G., Laub, M.T., Whitney, J.C.(2019) Nature 575: 674-678
- PubMed: 31695193
- DOI: https://doi.org/10.1038/s41586-019-1735-9
- Primary Citation of Related Structures:
6OTT, 6OX6 - PubMed Abstract:
Bacteria have evolved sophisticated mechanisms to inhibit the growth of competitors 1 . One such mechanism involves type VI secretion systems, which bacteria can use to inject antibacterial toxins directly into neighbouring cells. Many of these toxins target the integrity of the cell envelope, but the full range of growth inhibitory mechanisms remains unknown 2 . Here we identify a type VI secretion effector, Tas1, in the opportunistic pathogen Pseudomonas aeruginosa. The crystal structure of Tas1 shows that it is similar to enzymes that synthesize (p)ppGpp, a broadly conserved signalling molecule in bacteria that modulates cell growth rate, particularly in response to nutritional stress 3 . However, Tas1 does not synthesize (p)ppGpp; instead, it pyrophosphorylates adenosine nucleotides to produce (p)ppApp at rates of nearly 180,000 molecules per minute. Consequently, the delivery of Tas1 into competitor cells drives rapid accumulation of (p)ppApp, depletion of ATP, and widespread dysregulation of essential metabolic pathways, thereby resulting in target cell death. Our findings reveal a previously undescribed mechanism for interbacterial antagonism and demonstrate a physiological role for the metabolite (p)ppApp in bacteria.
Organizational Affiliation:
Michael DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, Ontario, Canada.