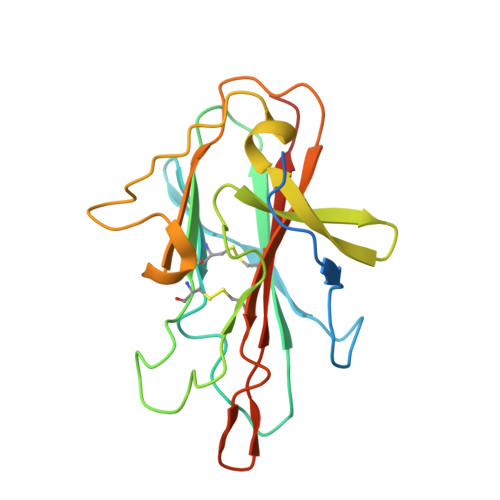
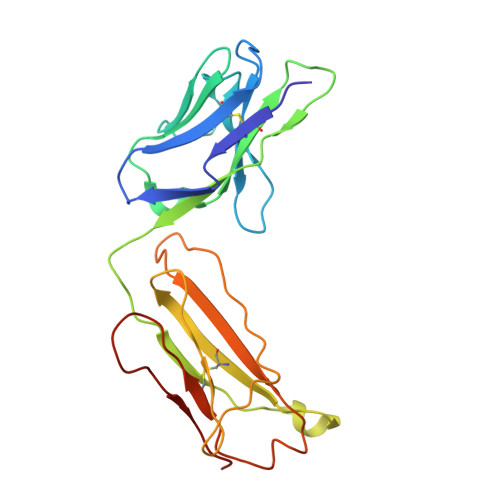
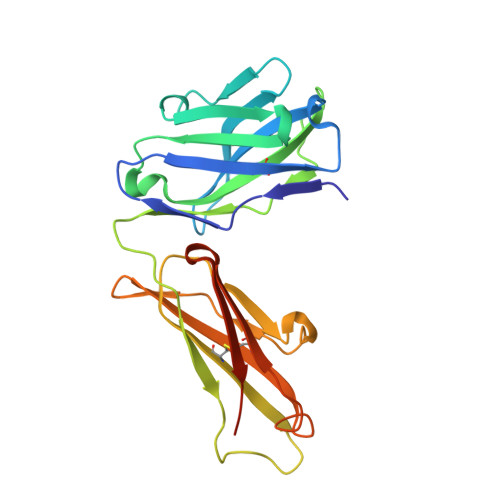
Structure and function of a malaria transmission blocking vaccine targeting Pfs230 and Pfs230-Pfs48/45 proteins.
Singh, K., Burkhardt, M., Nakuchima, S., Herrera, R., Muratova, O., Gittis, A.G., Kelnhofer, E., Reiter, K., Smelkinson, M., Veltri, D., Swihart, B.J., Shimp Jr., R., Nguyen, V., Zhang, B., MacDonald, N.J., Duffy, P.E., Garboczi, D.N., Narum, D.L.(2020) Commun Biol 3: 395-395
- PubMed: 32709983
- DOI: https://doi.org/10.1038/s42003-020-01123-9
- Primary Citation of Related Structures:
6OHG - PubMed Abstract:
Proteins Pfs230 and Pfs48/45 are Plasmodium falciparum transmission-blocking (TB) vaccine candidates that form a membrane-bound protein complex on gametes. The biological role of Pfs230 or the Pfs230-Pfs48/45 complex remains poorly understood. Here, we present the crystal structure of recombinant Pfs230 domain 1 (Pfs230D1M), a 6-cysteine domain, in complex with the Fab fragment of a TB monoclonal antibody (mAb) 4F12. We observed the arrangement of Pfs230 on the surface of macrogametes differed from that on microgametes, and that Pfs230, with no known membrane anchor, may exist on the membrane surface in the absence of Pfs48/45. 4F12 appears to sterically interfere with Pfs230 function. Combining mAbs against different epitopes of Pfs230D1 or of Pfs230D1 and Pfs48/45, significantly increased TB activity. These studies elucidate a mechanism of action of the Pfs230D1 vaccine, model the functional activity induced by a polyclonal antibody response and support the development of TB vaccines targeting Pfs230D1 and Pfs230D1-Pfs48/45.
Organizational Affiliation:
Structural Biology Section, Research Technologies Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 29 Lincoln Drive, Bethesda, MD, 20892, USA.