Engineering an anti-CD52 antibody for enhanced deamidation stability.
Qiu, H., Wei, R., Jaworski, J., Boudanova, E., Hughes, H., VanPatten, S., Lund, A., Day, J., Zhou, Y., McSherry, T., Pan, C.Q., Sendak, R.(2019) MAbs 11: 1266-1275
- PubMed: 31199181
- DOI: https://doi.org/10.1080/19420862.2019.1631117
- Primary Citation of Related Structures:
6OBD - PubMed Abstract:
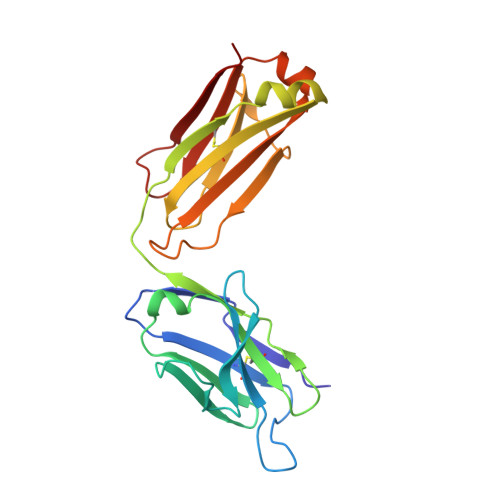
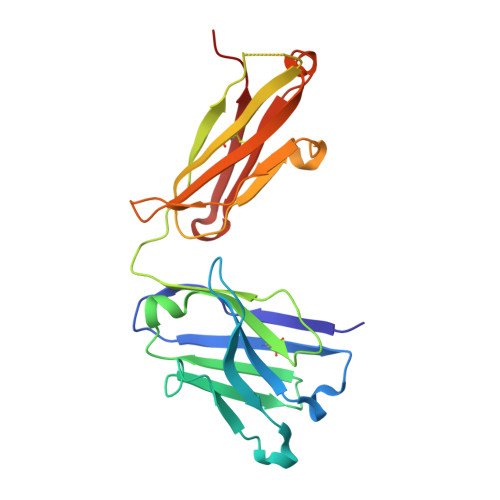
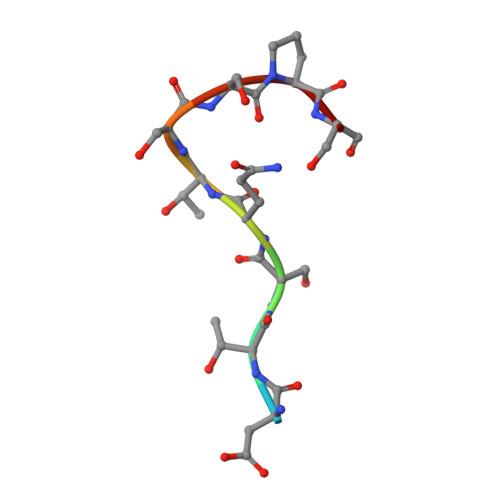
Deamidation evaluation and mitigation is an important aspect of therapeutic antibody developability assessment. We investigated the structure and function of the Asn-Gly deamidation in a human anti-CD52 IgG1 antibody light chain complementarity-determining region 1, and risk mitigation through protein engineering. Antigen binding affinity was found to decrease about 400-fold when Asn 33 was replaced with an Asp residue to mimic the deamidation product, suggesting significant impacts on antibody function. Other variants made at Asn 33 (N33H, N33Q, N33H, N33R) were also found to result in significant loss of antigen binding affinity. The co-crystal structure of the antigen-binding fragment bound to a CD52 peptide mimetic was solved at 2.2Å (PDB code 6OBD), which revealed that Asn 33 directly interacts with the CD52 phosphate group via a hydrogen bond. Gly 34 , but sits away from the binding interface, rendering it more amendable to mutagenesis without affecting affinity. Saturation mutants at Gly 34 were prepared and subjected to forced deamidation by incubation at elevated pH and temperature. Three mutants (G34R, G34K and G34Q) showed increased resistance to deamidation by LC-MS peptide mapping, while maintaining high binding affinity to CD52 antigen measured by Biacore. A complement -dependent cytotoxicity assay indicated that these mutants function by triggering antibody effector function. This study illustrates the importance of structure-based design and extensive mutagenesis to mitigate antibody developability issues.
Organizational Affiliation:
Biologics Research, Sanofi , Framingham , MA , USA.