A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites.
Koirala, D., Shao, Y., Koldobskaya, Y., Fuller, J.R., Watkins, A.M., Shelke, S.A., Pilipenko, E.V., Das, R., Rice, P.A., Piccirilli, J.A.(2019) Nat Commun 10: 3629-3629
- PubMed: 31399592
- DOI: https://doi.org/10.1038/s41467-019-11585-z
- Primary Citation of Related Structures:
6MWN - PubMed Abstract:
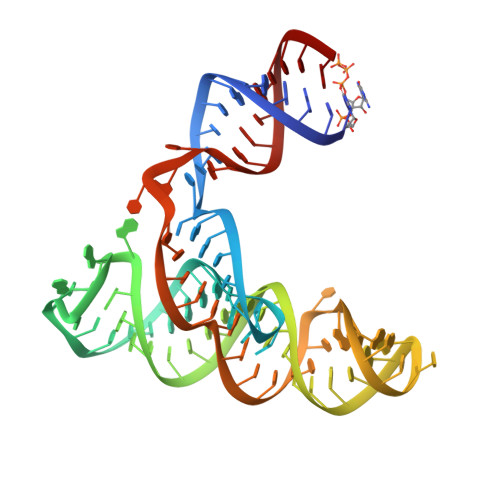
Picornaviral IRES elements are essential for initiating the cap-independent viral translation. However, three-dimensional structures of these elements remain elusive. Here, we report a 2.84-Å resolution crystal structure of hepatitis A virus IRES domain V (dV) in complex with a synthetic antibody fragment-a crystallization chaperone. The RNA adopts a three-way junction structure, topologically organized by an adenine-rich stem-loop motif. Despite no obvious sequence homology, the dV architecture shows a striking similarity to a circularly permuted form of encephalomyocarditis virus J-K domain, suggesting a conserved strategy for organizing the domain architecture. Recurrence of the motif led us to use homology modeling tools to compute a 3-dimensional structure of the corresponding domain of foot-and-mouth disease virus, revealing an analogous domain organizing motif. The topological conservation observed among these IRESs and other viral domains implicates a structured three-way junction as an architectural scaffold to pre-organize helical domains for recruiting the translation initiation machinery.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, 60637, USA.