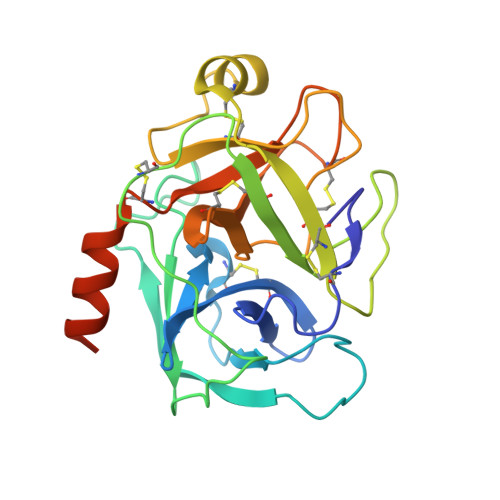
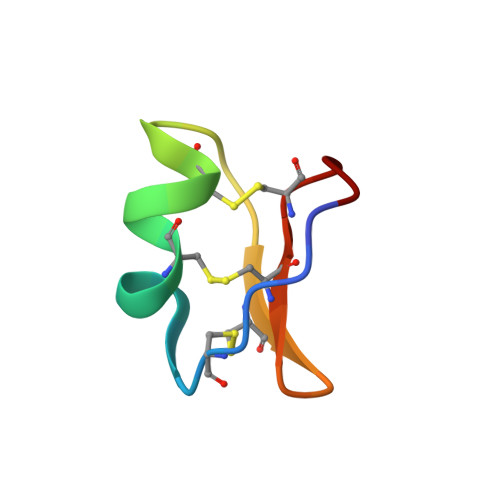
Head-to-Tail Cyclization after Interaction with Trypsin: A Scorpion Venom Peptide that Resembles Plant Cyclotides.
Mourao, C.B.F., Brand, G.D., Fernandes, J.P.C., Prates, M.V., Bloch Jr., C., Barbosa, J.A.R.G., Freitas, S.M., Restano-Cassulini, R., Possani, L.D., Schwartz, E.F.(2020) J Med Chem 63: 9500-9511
- PubMed: 32787139
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00686
- Primary Citation of Related Structures:
6MRQ - PubMed Abstract:
Peptidase inhibitors (PIs) have been broadly studied due to their wide therapeutic potential for human diseases. A potent trypsin inhibitor from Tityus obscurus scorpion venom was characterized and named ToPI1, with 33 amino acid residues and three disulfide bonds. The X-ray structure of the ToPI1:trypsin complex, in association with the mass spectrometry data, indicate a sequential set of events: the complex formation with the inhibitor Lys 32 in the trypsin S1 pocket, the inhibitor C-terminal residue Ser 33 cleavage, and the cyclization of ToPI1 via a peptide bond between residues Ile 1 and Lys 32 . Kinetic and thermodynamic characterization of the complex was obtained. ToPI1 shares no sequence similarity with other PIs characterized to date and is the first PI with CS-α/β motif described from animal venoms. In its cyclic form, it shares structural similarities with plant cyclotides that also inhibit trypsin. These results bring new insights for studies with venom compounds, PIs, and drug design.
Organizational Affiliation:
Neuropharma Lab, Departamento de Ciências Fisiológicas, Instituto de Ciências Biológicas, Universidade de Brası́lia, Brasília-DF 70910-900, Brazil.