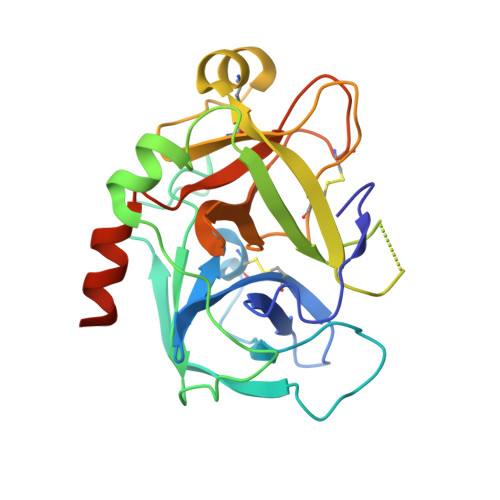
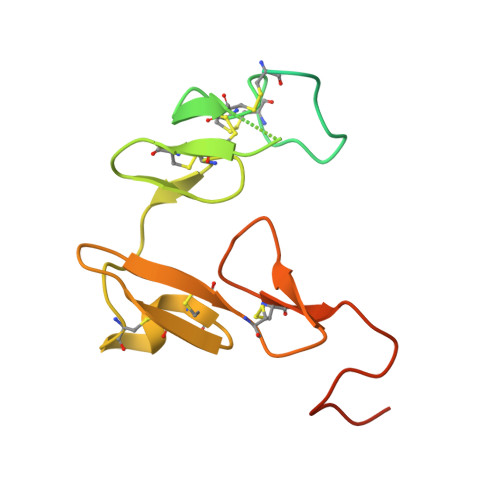
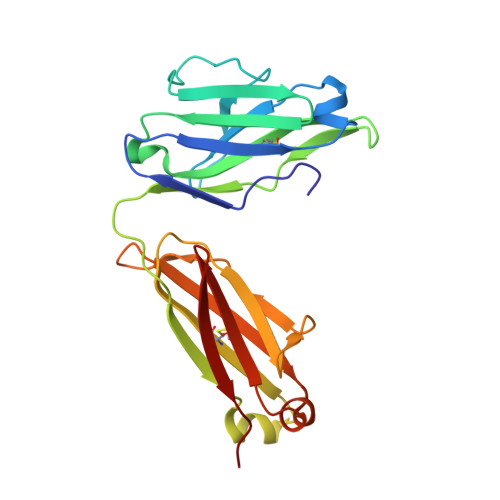
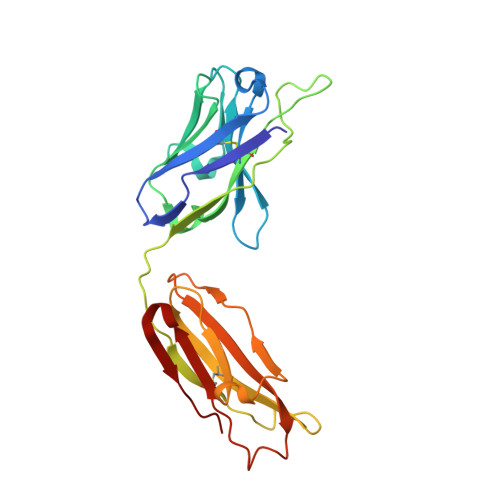
Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Zhao, X.Y., Wilmen, A., Wang, D., Wang, X., Bauzon, M., Kim, J.Y., Linden, L., Li, L., Egner, U., Marquardt, T., Moosmayer, D., Tebbe, J., Gluck, J.M., Ellinger, P., McLean, K., Yuan, S., Yegneswaran, S., Jiang, X., Evans, V., Gu, J.M., Schneider, D., Zhu, Y., Xu, Y., Mallari, C., Hesslein, A., Wang, Y., Schmidt, N., Gutberlet, K., Ruehl-Fehlert, C., Freyberger, A., Hermiston, T., Patel, C., Sim, D., Mosnier, L.O., Laux, V.(2020) Nat Commun 11: 2992-2992
- PubMed: 32532974
- DOI: https://doi.org/10.1038/s41467-020-16720-9
- Primary Citation of Related Structures:
6M3B, 6M3C - PubMed Abstract:
Activated protein C (APC) is a plasma serine protease with antithrombotic and cytoprotective functions. Based on the hypothesis that specific inhibition of APC's anticoagulant but not its cytoprotective activity can be beneficial for hemophilia therapy, 2 types of inhibitory monoclonal antibodies (mAbs) are tested: A type I active-site binding mAb and a type II mAb binding to an exosite on APC (required for anticoagulant activity) as shown by X-ray crystallography. Both mAbs increase thrombin generation and promote plasma clotting. Type I blocks all APC activities, whereas type II preserves APC's cytoprotective function. In normal monkeys, type I causes many adverse effects including animal death. In contrast, type II is well-tolerated in normal monkeys and shows both acute and prophylactic dose-dependent efficacy in hemophilic monkeys. Our data show that the type II mAb can specifically inhibit APC's anticoagulant function without compromising its cytoprotective function and offers superior therapeutic opportunities for hemophilia.
Organizational Affiliation:
US Innovation Center, Bayer, 455 Mission Bay Blvd. South, San Francisco, CA, 94158, USA. xiao-yan.zhao@bayer.com.