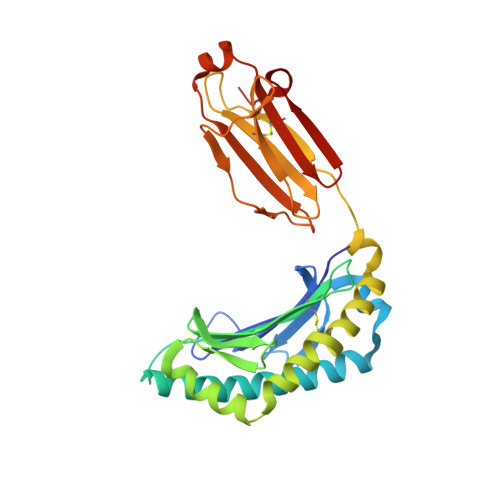
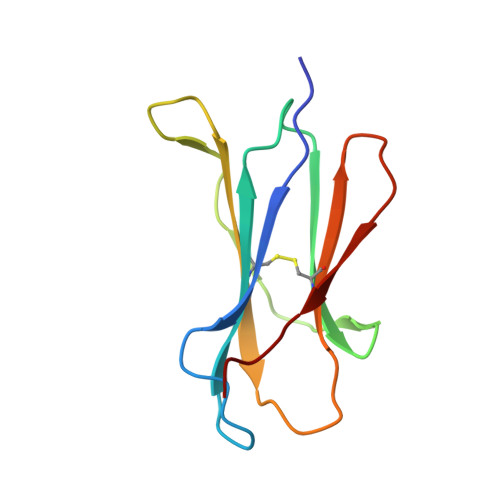
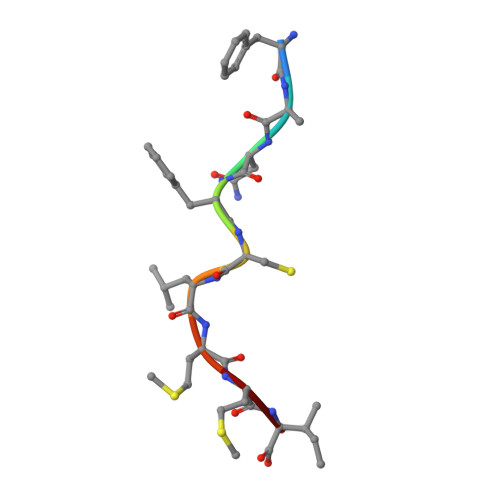
The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Li, Z., Zhang, N., Ma, L., Zhang, L., Meng, G., Xia, C.(2020) iScience 23: 101119-101119
- PubMed: 32438322
- DOI: https://doi.org/10.1016/j.isci.2020.101119
- Primary Citation of Related Structures:
5H5Z, 6LBE - PubMed Abstract:
Contemporary antigen presentation knowledge is based on the existence of a single β2m locus, and a classical MHC class I forms a complex with a peptide (i.e., pMHC-I) to trigger CTL immunity. However, two β2m loci have been found in diploid bony fish; the function of the two β2m molecules is unclear. Here, we determined the variant peptide profiles originating from different products of the β2m loci binding to the same MHC-I molecule and further solved the crystal structures of the two pMHC-I molecules (i.e., pCtid-UAA-β2m-2 and pCtid-UAA-β2m-1-II). Of note, in pCtid-UAA-β2m-2, a unique hydrogen bond network formed in the bottom of the peptide-binding groove (PBG) led to α2-helix drift, ultimately leading to structural changes in the PBG. The mechanism of the change in peptide presentation profiles by β2m molecules is illustrated. The results are also of great significance for antivirus and antitumor functions in cold-blooded vertebrates and even humans.
Organizational Affiliation:
Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing, 100193, China.