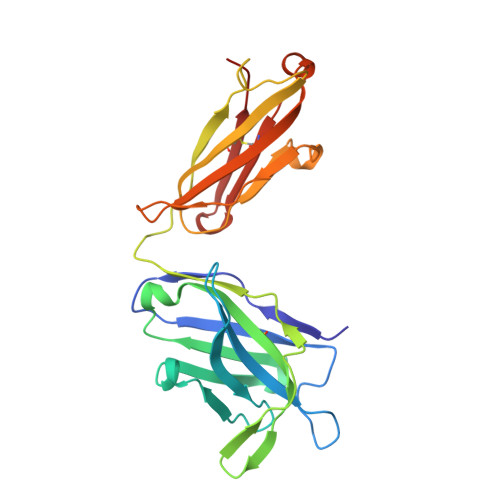
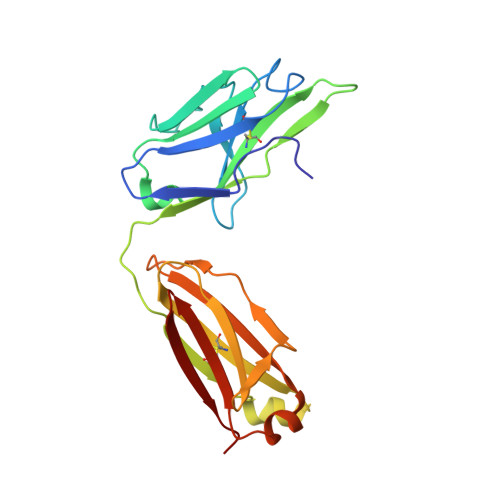
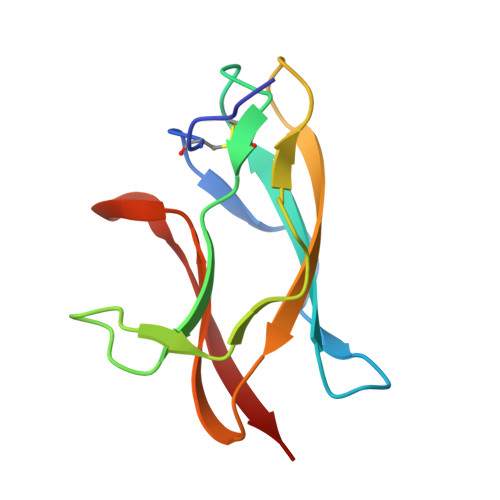Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Wang, L., Wang, R., Wang, L., Ben, H., Yu, L., Gao, F., Shi, X., Yin, C., Zhang, F., Xiang, Y., Zhang, L.(2019) Cell Rep 26: 3360-3368.e5
- PubMed: 30893607
- DOI: https://doi.org/10.1016/j.celrep.2019.02.062
- Primary Citation of Related Structures:
6JEP, 6JFH, 6JFI - PubMed Abstract:
We previously reported a human monoclonal antibody, ZK2B10, capable of protection against Zika virus (ZIKV) infection and microcephaly in developing mouse embryos. Here, we report the structural features and mechanism of action of ZK2B10. The crystal structure at a resolution of 2.32 Å revealed that the epitope is located on the lateral ridge of DIII of the envelope glycoprotein. Cryo-EM structure with mature ZIKV showed that the antibody binds to DIIIs around the icosahedral 2-fold, 3-fold, and 5-fold axes, a distinct feature compared to those reported for DIII-specific antibodies. The binding of ZK2B10 to ZIKV has no detectable effect on viral attachment to target cells or on conformational changes of the E glycoprotein in the acidic environment, suggesting that ZK2B10 functions at steps between the formation of the fusion intermediate and membrane fusion. These results provide structural and mechanistic insights into how ZK2B10 mediates protection against ZIKV infection.
Organizational Affiliation:
Beijing Advanced Innovation Center for Structural Biology, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Center for Global Health and Infectious Diseases, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.