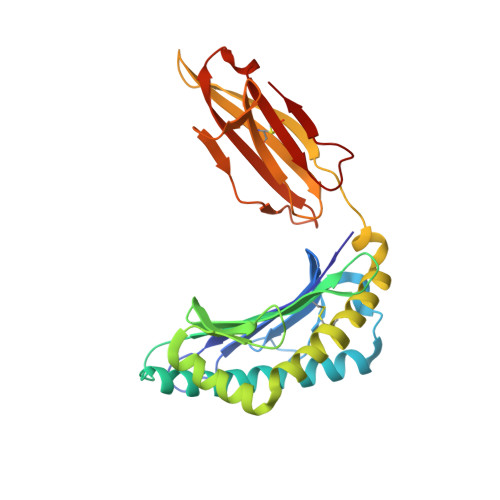
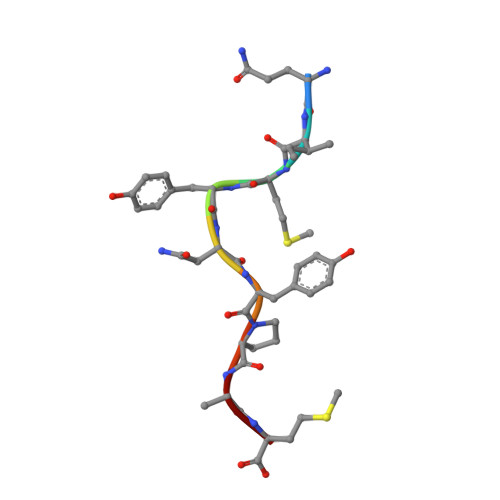
Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Zhu, S., Liu, K., Chai, Y., Wu, Y., Lu, D., Xiao, W., Cheng, H., Zhao, Y., Ding, C., Lyu, J., Lou, Y., Gao, G.F., Liu, W.J.(2019) Front Immunol 10: 1709-1709
- PubMed: 31396224
- DOI: https://doi.org/10.3389/fimmu.2019.01709
- Primary Citation of Related Structures:
6J1V, 6J1W, 6J29, 6J2A - PubMed Abstract:
Human leukocyte antigen (HLA) alleles have a high degree of polymorphism, which determines their peptide-binding motifs and subsequent T-cell receptor recognition. The simplest way to understand the cross-presentation of peptides by different alleles is to classify these alleles into supertypes. A1 and A3 HLA supertypes are widely distributed in humans. However, direct structural and functional evidence for peptide presentation features of key alleles (e.g., HLA-A * 30:01 and -A * 30:03) are lacking. Herein, the molecular basis of peptide presentation of HLA-A * 30:01 and -A * 30:03 was demonstrated by crystal structure determination and thermostability measurements of complexes with T-cell epitopes from influenza virus (NP44), human immunodeficiency virus (RT313), and Mycobacterium tuberculosis (MTB). When binding to the HIV peptide, RT313, the PΩ-Lys anchoring modes of HLA-A * 30:01, and -A * 30:03 were similar to those of HLA-A * 11:01 in the A3 supertype. However, HLA-A * 30:03, but not -A * 30:01, also showed binding with the HLA * 01:01-favored peptide, NP44, but with a specific structural conformation. Thus, different from our previous understanding, HLA-A * 30:01 and -A * 30:03 have specific peptide-binding characteristics that may lead to their distinct supertype-featured binding peptide motifs. Moreover, we also found that residue 77 in the F pocket was one of the key residues for the divergent peptide presentation characteristics of HLA-A * 30:01 and -A * 30:03. Interchanging residue 77 between HLA-A * 30:01 and HLA-A * 30:03 switched their presented peptide profiles. Our results provide important recommendations for screening virus and tumor-specific peptides among the population with prevalent HLA supertypes for vaccine development and immune interventions.
Organizational Affiliation:
School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China.