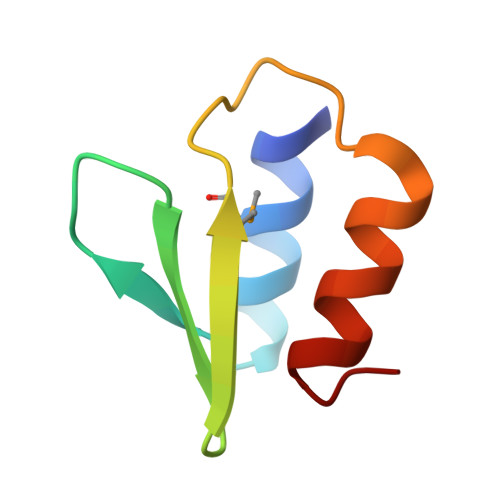
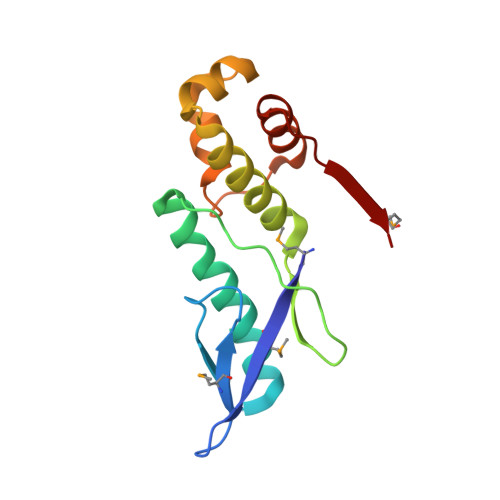
The E. coli HicB Antitoxin Contains a Structurally Stable Helix-Turn-Helix DNA Binding Domain.
Manav, M.C., Turnbull, K.J., Jurenas, D., Garcia-Pino, A., Gerdes, K., Brodersen, D.E.(2019) Structure 27: 1675-1685.e3
- PubMed: 31495532
- DOI: https://doi.org/10.1016/j.str.2019.08.008
- Primary Citation of Related Structures:
6HPB, 6HPC - PubMed Abstract:
The E. coli hicAB type II toxin-antitoxin locus is unusual by being controlled by two promoters and by having the toxin encoded upstream of the antitoxin. HicA toxins contain a double-stranded RNA-binding fold and cleaves both mRNA and tmRNA in vivo, while HicB antitoxins contain a partial RNase H fold and either a helix-turn-helix (HTH) or ribbon-helix-helix domain. It is not known how an HTH DNA-binding domain affects higher-order structure for the HicAB modules. Here, we present crystal structures of the isolated E. coli HicB antitoxin and full-length HicAB complex showing that HicB forms a stable DNA-binding module and interacts in a canonical way with HicA despite the presence of an HTH-type DNA-binding domain. No major structural rearrangements take place upon binding of the toxin. Both structures expose well-ordered DNA-binding motifs allowing a model for DNA binding by the antitoxin to be generated.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Centre for Bacterial Stress Response and Persistence, Aarhus 8000, Denmark.