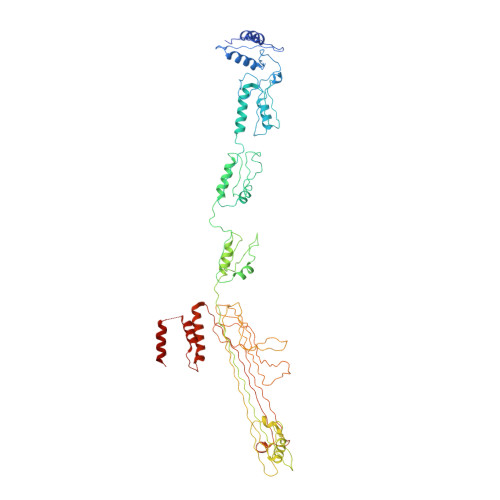
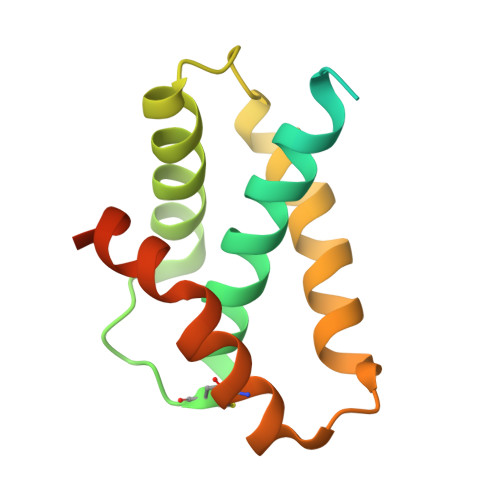
Core architecture of a bacterial type II secretion system.
Chernyatina, A.A., Low, H.H.(2019) Nat Commun 10: 5437-5437
- PubMed: 31780649
- DOI: https://doi.org/10.1038/s41467-019-13301-3
- Primary Citation of Related Structures:
6HCG - PubMed Abstract:
Bacterial type II secretion systems (T2SSs) translocate virulence factors, toxins and enzymes across the cell outer membrane. Here we use negative stain and cryo-electron microscopy to reveal the core architecture of an assembled T2SS from the pathogen Klebsiella pneumoniae. We show that 7 proteins form a ~2.4 MDa complex that spans the cell envelope. The outer membrane complex includes the secretin PulD, with all domains modelled, and the pilotin PulS. The inner membrane assembly platform components PulC, PulE, PulL, PulM and PulN have a relative stoichiometric ratio of 2:1:1:1:1. The PulE ATPase, PulL and PulM combine to form a flexible hexameric hub. Symmetry mismatch between the outer membrane complex and assembly platform is overcome by PulC linkers spanning the periplasm, with PulC HR domains binding independently at the secretin base. Our results show that the T2SS has a highly dynamic modular architecture, with implication for pseudo-pilus assembly and substrate loading.
Organizational Affiliation:
Department of Life Sciences, Imperial College, London, SW7 2AZ, UK.