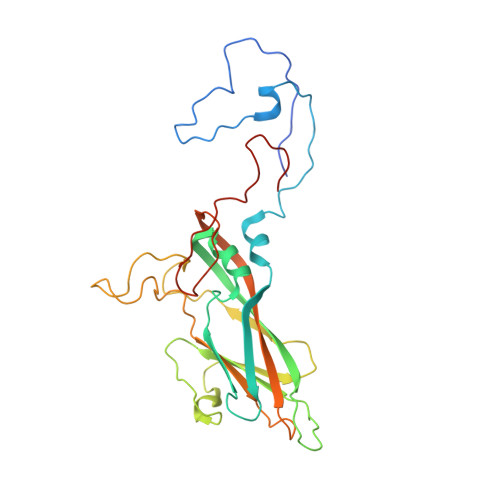
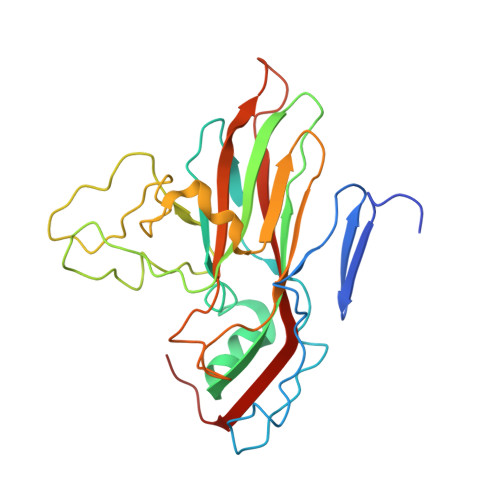
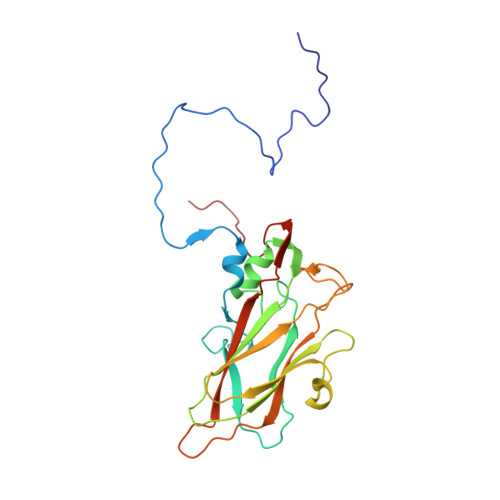
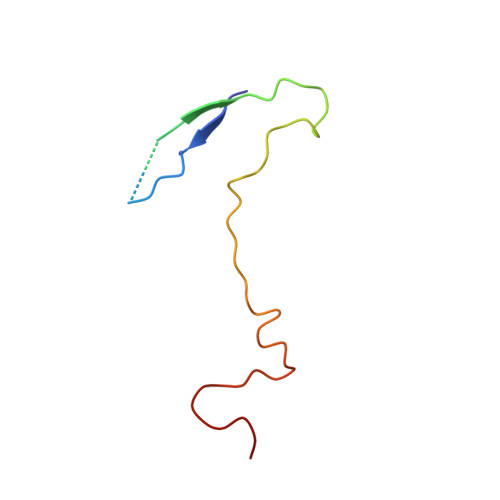
A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses.
Abdelnabi, R., Geraets, J.A., Ma, Y., Mirabelli, C., Flatt, J.W., Domanska, A., Delang, L., Jochmans, D., Kumar, T.A., Jayaprakash, V., Sinha, B.N., Leyssen, P., Butcher, S.J., Neyts, J.(2019) PLoS Biol 17: e3000281-e3000281
- PubMed: 31185007
- DOI: https://doi.org/10.1371/journal.pbio.3000281
- Primary Citation of Related Structures:
6GZV - PubMed Abstract:
Rhino- and enteroviruses are important human pathogens, against which no antivirals are available. The best-studied inhibitors are "capsid binders" that fit in a hydrophobic pocket of the viral capsid. Employing a new class of entero-/rhinovirus inhibitors and by means of cryo-electron microscopy (EM), followed by resistance selection and reverse genetics, we discovered a hitherto unknown druggable pocket that is formed by viral proteins VP1 and VP3 and that is conserved across entero-/rhinovirus species. We propose that these inhibitors stabilize a key region of the virion, thereby preventing the conformational expansion needed for viral RNA release. A medicinal chemistry effort resulted in the identification of analogues targeting this pocket with broad-spectrum activity against Coxsackieviruses B (CVBs) and compounds with activity against enteroviruses (EV) of groups C and D, and even rhinoviruses (RV). Our findings provide novel insights in the biology of the entry of entero-/rhinoviruses and open new avenues for the design of broad-spectrum antivirals against these pathogens.
Organizational Affiliation:
Department of Microbiology and Immunology, Rega Institute for Medical Research, Laboratory of Virology and Chemotherapy, KU Leuven, Leuven, Belgium.