The putative polysaccharide deacetylase Ba0331: cloning, expression, crystallization and structure determination.
Andreou, A., Giastas, P., Arnaouteli, S., Tzanodaskalaki, M., Tzartos, S.J., Bethanis, K., Bouriotis, V., Eliopoulos, E.E.(2019) Acta Crystallogr F Struct Biol Commun 75: 312-320
- PubMed: 30950833
- DOI: https://doi.org/10.1107/S2053230X19001766
- Primary Citation of Related Structures:
6GO1 - PubMed Abstract:
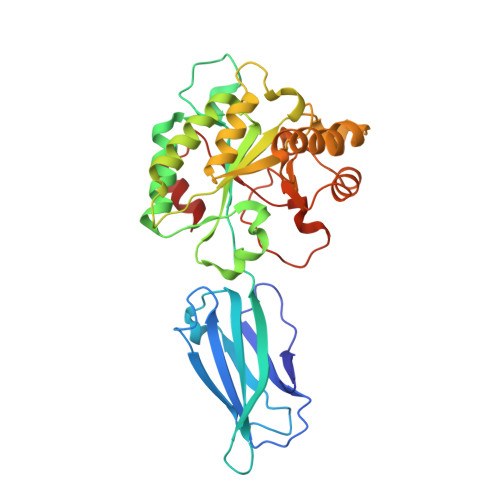
Ba0331 is a putative polysaccharide deacetylase from Bacillus anthracis, the etiological agent of the disease anthrax, that contributes to adaptation of the bacterium under extreme conditions and to maintenance of the cell shape. In the present study, the crystal structure of Ba0331 was determined at 2.6 Å resolution. The structure consists of two domains: a fibronectin type 3-like (Fn3-like) domain and a NodB catalytic domain. The latter is present in all carbohydrate esterase family 4 enzymes, while a comparative analysis of the Fn3-like domain revealed structural plasticity despite the retention of the conserved Fn3-like domain characteristics.
Organizational Affiliation:
Department of Biotechnology, Agricultural University of Athens, Iera Odos 75, 11855 Athens, Greece.