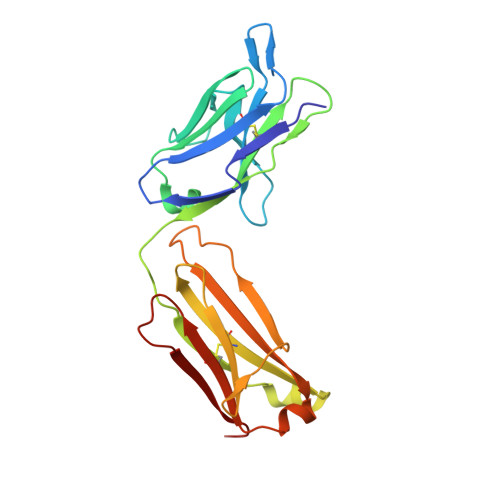
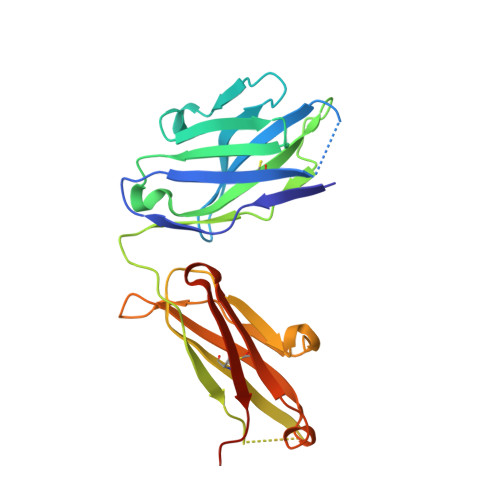
Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Chen, J.B., Ramadani, F., Pang, M.O.Y., Beavil, R.L., Holdom, M.D., Mitropoulou, A.N., Beavil, A.J., Gould, H.J., Chang, T.W., Sutton, B.J., McDonnell, J.M., Davies, A.M.(2018) Sci Rep 8: 11548-11548
- PubMed: 30069035
- DOI: https://doi.org/10.1038/s41598-018-29664-4
- Primary Citation of Related Structures:
6EYN - PubMed Abstract:
Immunoglobulin E (IgE) antibodies play a central role in the allergic response: interaction with FcεRI on mast cells and basophils leads to immediate hypersensitivity reactions upon allergen challenge, while interaction with CD23/FcεRII, expressed on a variety of cells, regulates IgE synthesis among other activities. The receptor-binding IgE-Fc region has recently been found to display remarkable flexibility, from acutely bent to extended conformations, with allosteric communication between the distant FcεRI and CD23 binding sites. We report the structure of an anti-IgE antibody Fab (8D6) bound to IgE-Fc through a mixed protein-carbohydrate epitope, revealing further flexibility and a novel extended conformation with potential relevance to that of membrane-bound IgE in the B cell receptor for antigen. Unlike the earlier, clinically approved anti-IgE antibody omalizumab, 8D6 inhibits binding to FcεRI but not CD23; the structure reveals how this discrimination is achieved through both orthosteric and allosteric mechanisms, supporting therapeutic strategies that retain the benefits of CD23 binding.
Organizational Affiliation:
Genomics Research Center, Academia Sinica, Taipei, 115, Taiwan.