6ENY
Structure of the human PLC editing module
- PDB DOI: https://doi.org/10.2210/pdb6ENY/pdb
- EM Map EMD-3906: EMDB EMDataResource
- Classification: IMMUNE SYSTEM
- Organism(s): Homo sapiens
- Mutation(s): No
- Deposited: 2017-10-07 Released: 2017-11-29
- Funding Organization(s): German Research Foundation
Experimental Data Snapshot
- Method: ELECTRON MICROSCOPY
- Resolution: 5.80 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
wwPDB Validation 3D Report Full Report
This is version 2.0 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | A [auth B] | 99 | Homo sapiens | Mutation(s): 0 | 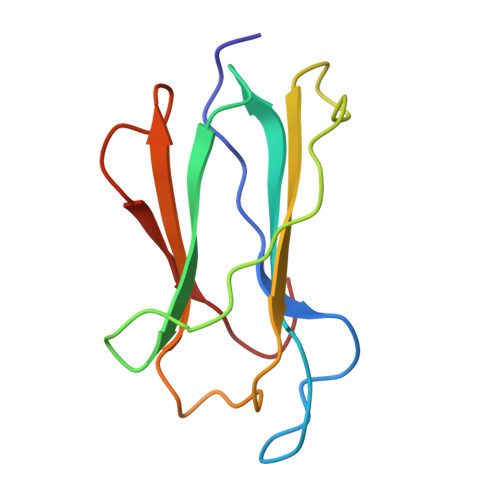 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tapasin | B [auth C] | 428 | Homo sapiens | Mutation(s): 0 | 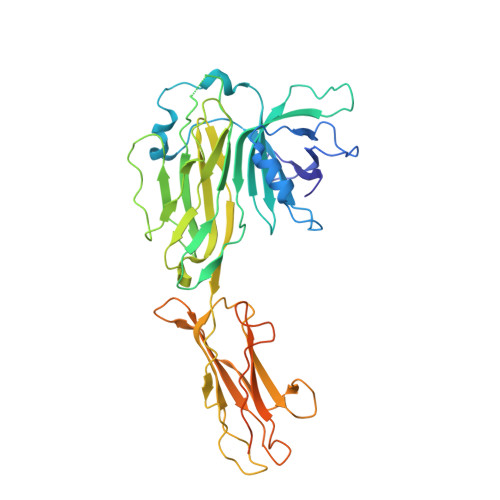 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O15533 (Homo sapiens) Explore O15533 Go to UniProtKB: O15533 | |||||
PHAROS: O15533 GTEx: ENSG00000231925 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O15533 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein disulfide-isomerase A3 | C [auth D] | 481 | Homo sapiens | Mutation(s): 0 EC: 5.3.4.1 | 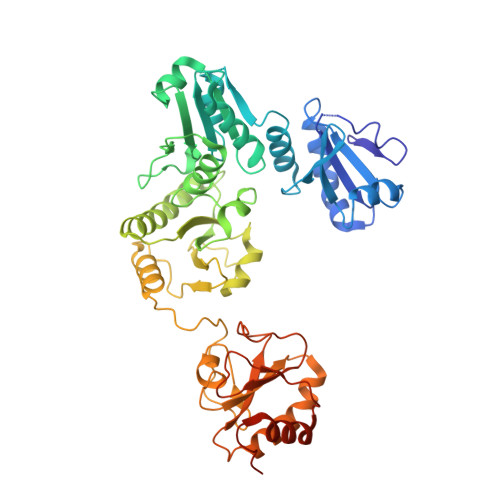 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30101 (Homo sapiens) Explore P30101 Go to UniProtKB: P30101 | |||||
PHAROS: P30101 GTEx: ENSG00000167004 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30101 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HLA class I histocompatibility antigen, A-3 alpha chain | D [auth F] | 341 | Homo sapiens | Mutation(s): 0 | 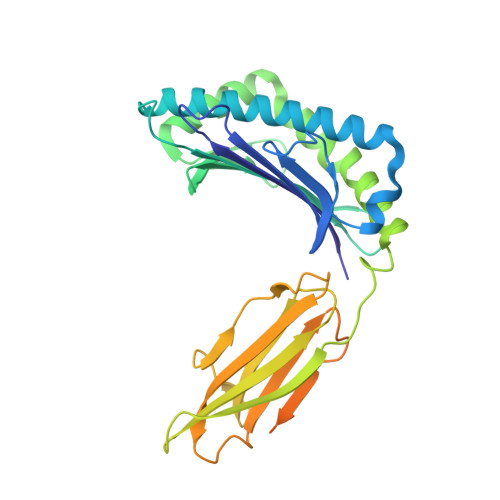 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04439 (Homo sapiens) Explore P04439 Go to UniProtKB: P04439 | |||||
PHAROS: P04439 GTEx: ENSG00000206503 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04439 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calreticulin | E [auth G] | 400 | Homo sapiens | Mutation(s): 0 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P27797 (Homo sapiens) Explore P27797 Go to UniProtKB: P27797 | |||||
PHAROS: P27797 GTEx: ENSG00000179218 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27797 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | F [auth A] | 2 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | G [auth E] | 4 | N/A | N/A | |
Experimental Data & Validation
Experimental Data
- Method: ELECTRON MICROSCOPY
- Resolution: 5.80 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | FREALIGN | X |
| MODEL REFINEMENT | PHENIX |
Entry History & Funding Information
Deposition Data
- Released Date: 2017-11-29 Deposition Author(s): Trowitzsch, S., Januliene, D., Blees, A., Moeller, A., Tampe, R.
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation | Germany | SFB 807 |
| German Research Foundation | Germany | GRK 1986 |
Revision History (Full details and data files)
- Version 1.0: 2017-11-29
Type: Initial release - Version 1.1: 2017-12-06
Changes: Database references - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Advisory, Atomic model, Data collection, Derived calculations, Structure summary













