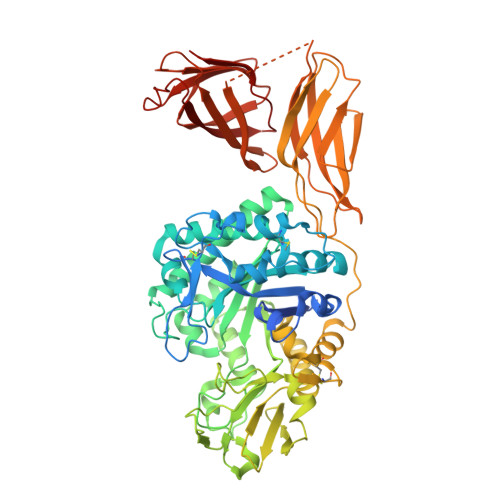
The crystal structure of the chitinase ChiA74 of Bacillus thuringiensis has a multidomain assembly.
Juarez-Hernandez, E.O., Casados-Vazquez, L.E., Brieba, L.G., Torres-Larios, A., Jimenez-Sandoval, P., Barboza-Corona, J.E.(2019) Sci Rep 9: 2591-2591
- PubMed: 30796308
- DOI: https://doi.org/10.1038/s41598-019-39464-z
- Primary Citation of Related Structures:
6BT9 - PubMed Abstract:
There is no structural information about any chitinase synthesized by Bacillus thuringiensis, the most successful microbial insect larvicide used worldwide. In this study, we solved the 3D structure of the chitinase ChiA74 at 2.26 Å. The crystal structure shows that ChiA74 is composed of a modular arrangement formed by (i) a catalytic region (CD), (ii) a chitinase insertion domain (CID), (iii) a fibronectin type III domain (FnIII), and (iv) a chitin binding domain (CBD). The location of the CBD with respect to the CD has no structural similarity to other chitinases with known structures. The activity of a ChiA74 lacking its secretion signal peptide (ChiA74Δsp) and a truncated version lacking its CBD/FnIII domains (ChiA74Δsp-50) did not have statistical differences in activity against colloidal chitin. However, ChiA74Δsp exhibits 4.5 and 2.0 higher activity than versions lacking the CBD (ChiA74Δsp-60) and CBD/FnIII domains (ChiA74Δsp-50), respectively, when crystalline chitin was used as substrate. Our data suggest that the CBD might plays a significant role in crystalline chitin hydrolysis. We also demonstrated the importance of the catalytic E211 in the CD, as mutants ChiA74Δsp E211N and ChiA74Δsp D207N, E211N were inactive against colloidal and crystalline chitins, chitosan and 4-MU-GlcNAc 3 . ChiA74 has a processive activity producing oligosaccharides with degree of polymerization (DP) of 1 (GlcNAc) and 2 (GlcNAc 2 ).
Organizational Affiliation:
Universidad de Guanajuato Campus Irapuato-Salamanca, División de Ciencias de la Vida, Posgrado en Biociencias, Irapuato, Guanajuato, 36500, Mexico.