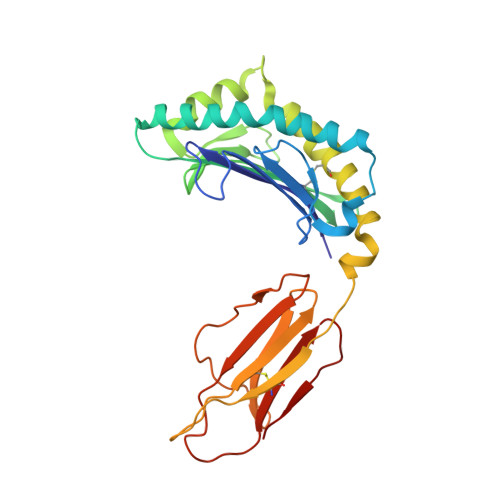
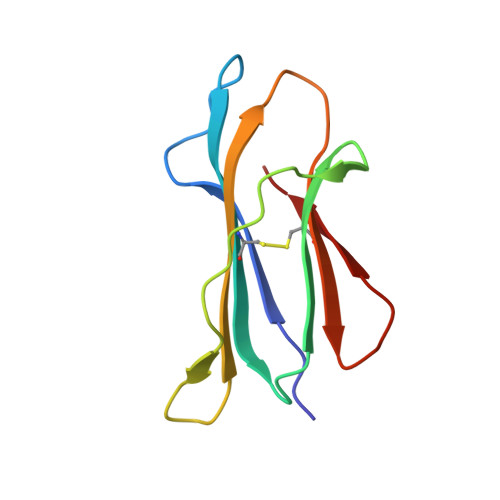
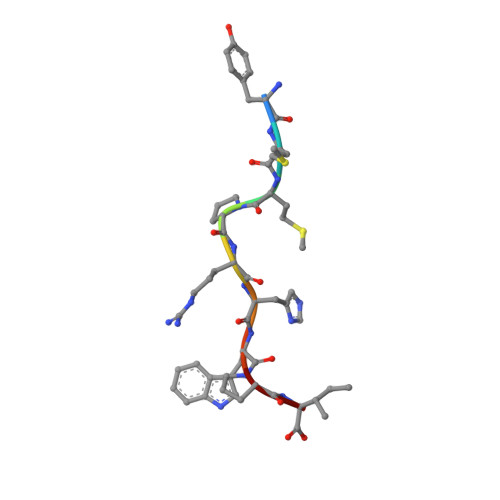
A Glimpse of the Peptide Profile Presentation byXenopus laevisMHC Class I: Crystal Structure of pXela-UAA Reveals a Distinct Peptide-Binding Groove.
Ma, L., Zhang, N., Qu, Z., Liang, R., Zhang, L., Zhang, B., Meng, G., Dijkstra, J.M., Li, S., Xia, M.C.(2020) J Immunol 204: 147-158
- PubMed: 31776204
- DOI: https://doi.org/10.4049/jimmunol.1900865
- Primary Citation of Related Structures:
6A2B - PubMed Abstract:
The African clawed frog, Xenopus laevis , is a model species for amphibians. Before metamorphosis, tadpoles do not efficiently express the single classical MHC class I (MHC-I) molecule Xela -UAA, but after metamorphosis, adults express this molecule in abundance. To elucidate the Ag-presenting mechanism of Xela -UAA, in this study, the Xela -UAA structure complex (p Xela -UAAg) bound with a peptide from a synthetic random peptide library was determined. The amino acid homology between the Xela -UAA and MHC-I sequences of different species is <45%, and these differences are fully reflected in the three-dimensional structure of p Xela -UAAg. Because of polymorphisms and interspecific differences in amino acid sequences, p Xela -UAAg forms a distinct peptide-binding groove and presents a unique peptide profile. The most important feature of p Xela -UAAg is the two-amino acid insertion in the α2-helical region, which forms a protrusion of ∼3.8 Å that is involved in TCR docking. Comparison of peptide-MHC-I complex (pMHC-I) structures showed that only four amino acids in β2-microglobulin that were bound to MHC-I are conserved in almost all jawed vertebrates, and the most unique feature in nonmammalian pMHC-I molecules is that the AB loop bound β2-microglobulin. Additionally, the binding distance between pMHC-I and CD8 molecules in nonmammals is different from that in mammals. These unique features of p Xela -UAAg provide enhanced knowledge of T cell immunity and bridge the knowledge gap regarding the coevolutionary progression of the MHC-I complex from aquatic to terrestrial species.
Organizational Affiliation:
Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing 100193, China.