Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Li, Z., Liu, H., Li, J., Yang, Q., Feng, Z., Li, Y., Yang, H., Yu, C., Wan, J., Liu, W., Zhang, M.(2019) Structure 27: 27-38.e4
- PubMed: 30503778
- DOI: https://doi.org/10.1016/j.str.2018.10.011
- Primary Citation of Related Structures:
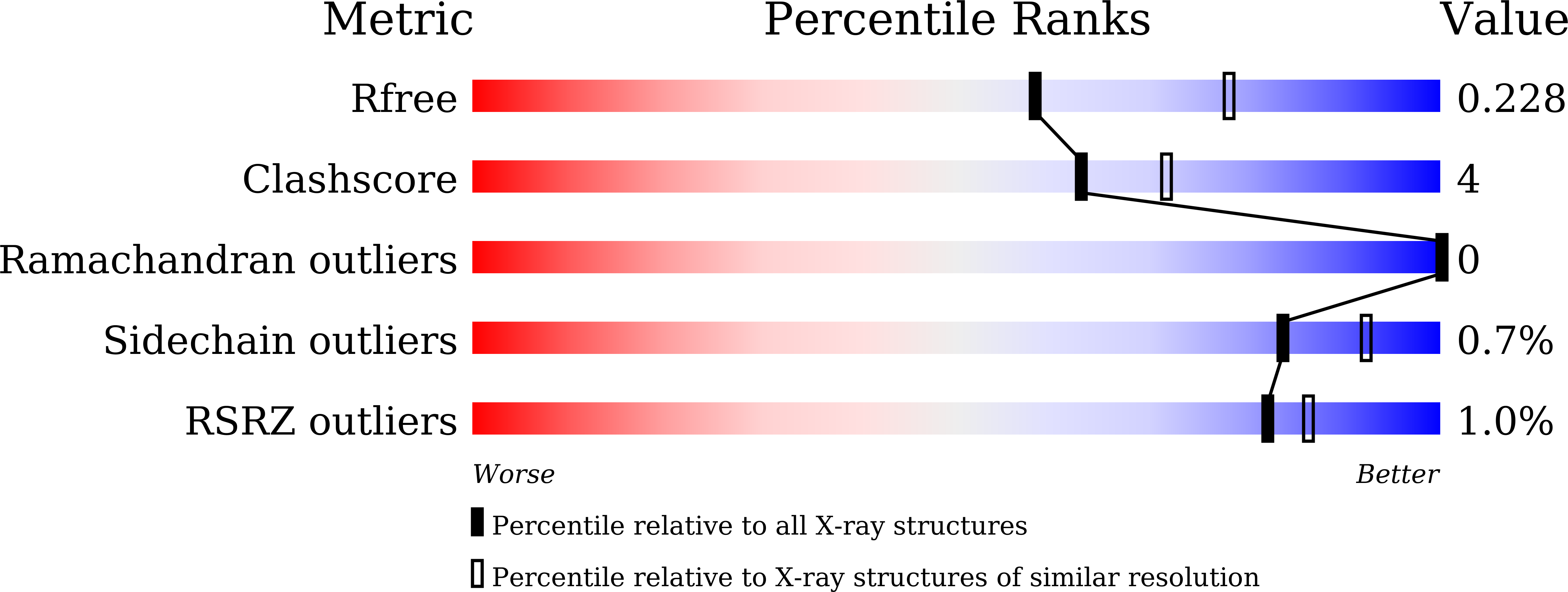
5ZZ9 - PubMed Abstract:
Drebrin is an actin bundling protein that plays critical roles in synaptic spine development and plasticity. Homer, one of the most abundant scaffolding proteins in postsynaptic density, interacts with Drebrin's C-terminal PPXXF motifs using its Ena/VASP homology 1 (EVH1) domain. However, the molecular mechanism and biological function of this interaction remain unclear. Here we show that Homer specifically binds to the first but not the second PPXXF motif in Drebrin. The crystal structure of Drebrin-Homer binding motif 1 in complex with Homer EVH1 reveals a consensus Homer EVH1 binding motif. Homer tetramer promotes actin bundling activity of Drebrin in vitro and stimulates Drebrin-induced filopodia formation and elongation in cells. We further show that monomeric Homer1a antagonizes Homer1b in promoting Drebrin-stimulated actin bundling. Our study suggests a potential regulatory role of Homer1 in modulating excitatory synaptic spine homeostatic scaling via binding to Drebrin.
Organizational Affiliation:
Shenzhen Key Laboratory for Neuronal Structural Biology, Biomedical Research Institute, Shenzhen Peking University-The Hong Kong University of Science and Technology Medical Center, Shenzhen 518036, China.