The C-terminal end of mycobacterial HadBC regulates AcpM interaction during the FAS-II pathway: a structural perspective.
Singh, B.K., Biswas, R., Bhattacharyya, S., Basak, A., Das, A.K.(2022) FEBS J
- PubMed: 35175661
- DOI: https://doi.org/10.1111/febs.16405
- Primary Citation of Related Structures:
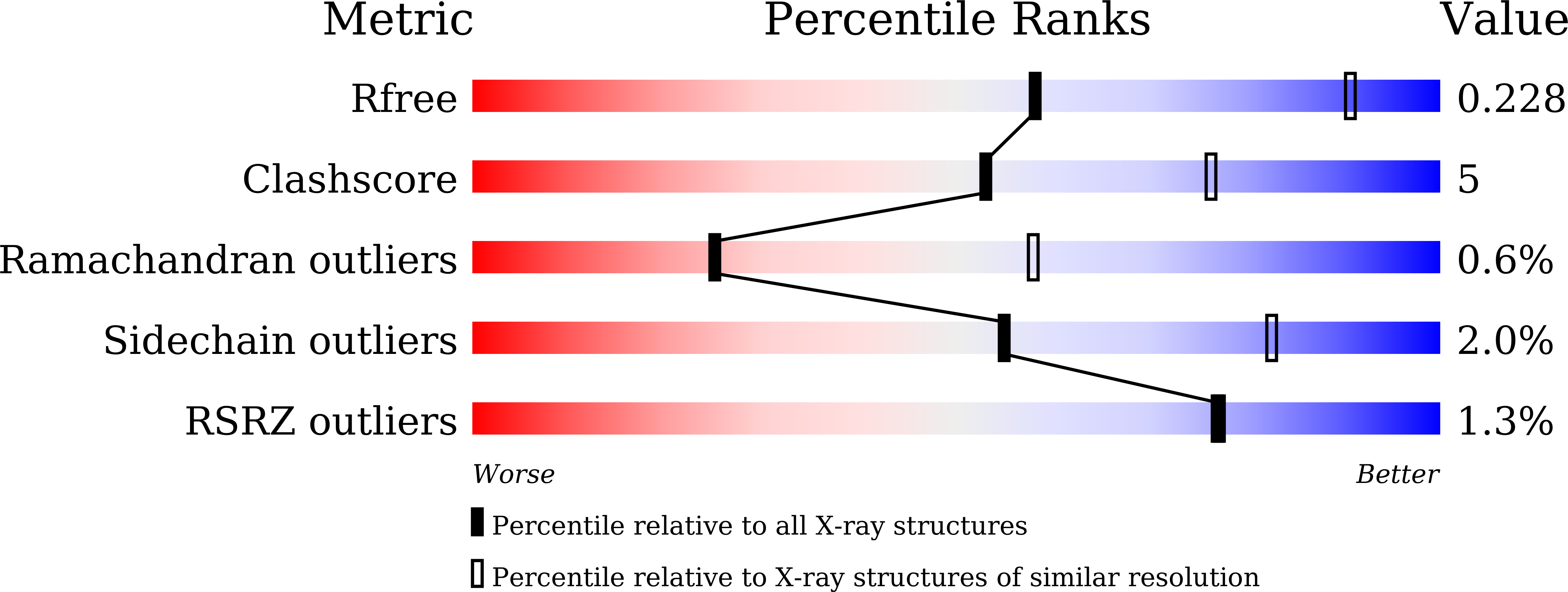
5ZY8 - PubMed Abstract:
Comprehending the molecular strategies employed by Mycobacterium tuberculosis (Mtb) in FAS-II regulation is of paramount significance for curbing tuberculosis progression. Mtb employs two sets of dehydratases, namely HadAB and HadBC (β-hydroxyacyl acyl carrier protein dehydratase), for the regulation of the fatty acid synthase (FAS-II) pathway. We utilized a sequence similarity network to discern the basis for the presence of two copies of the dehydratase gene in Mtb. This analysis groups HadC and HadA in different clusters, which could be attributed to the variability in their physiological role with respect to the acyl chain uptake. Our study reveals structural details pertaining to the crystal structure of the last remaining enzyme of the FAS-II pathway. It also provides insights into the highly flexible hot-dog helix and substrate regulatory loop. Additionally, mutational studies assisted in establishing the role of the C-terminal end in HadC of HadBC in the regulation of acyl carrier protein from Mtb-mediated interactions. Complemented with surface plasmon resonance and molecular dynamics simulation studies, the present study provides the first evidence of the molecular mechanisms involved in the differential binding affinity of the acyl carrier protein from Mtb towards both mtbHadAB and mtbHadBC.
Organizational Affiliation:
School of Biosciences, Indian Institute of Technology Kharagpur, India.