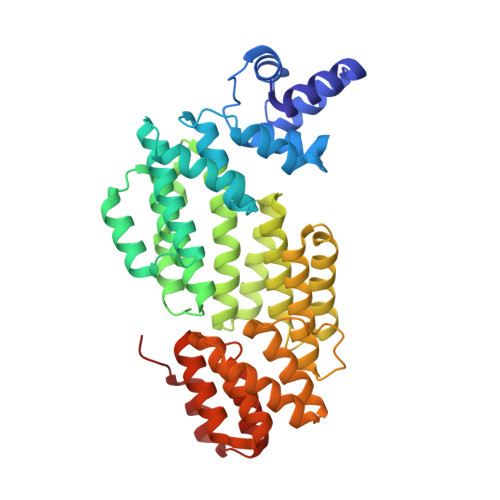
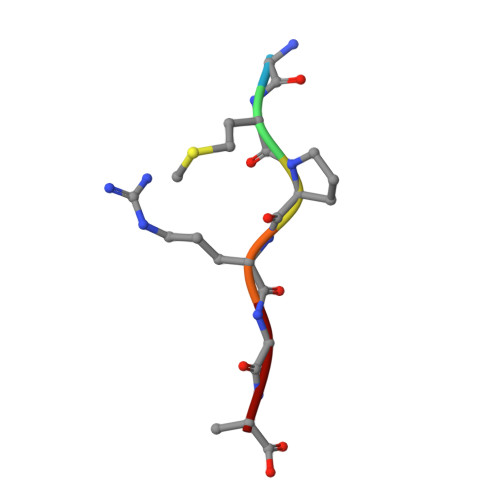
Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Dou, C., Xiong, J., Gu, Y., Yin, K., Wang, J., Hu, Y., Zhou, D., Fu, X., Qi, S., Zhu, X., Yao, S., Xu, H., Nie, C., Liang, Z., Yang, S., Wei, Y., Cheng, W.(2018) Nat Microbiol 3: 1285-1294
- PubMed: 30323253
- DOI: https://doi.org/10.1038/s41564-018-0259-7
- Primary Citation of Related Structures:
5ZVV, 5ZVW, 5ZW5, 5ZW6 - PubMed Abstract:
Communication is vital for all organisms including microorganisms, which is clearly demonstrated by the bacterial quorum-sensing system. However, the molecular mechanisms underlying communication among viruses (phages) via the quorum-sensing-like 'arbitrium' system remain unclear. Viral or host densities are known to be related to an increased prevalence of lysogeny; however, how the switch from the lytic to the lysogenic pathway occurs is unknown. Thus, we sought to reveal mechanisms of communication among viruses and determine the lysogenic dynamics involved. Structural and functional analyses of the phage-derived SAIRGA and GMPRGA peptides and their corresponding receptors, phAimR and spAimR, indicated that SAIRGA directs the lysis-lysogeny decision of phi3T by modulating conformational changes in phAimR, whereas GMPRGA regulates the lysis-lysogeny pathway by stabilizing spAimR in the dimeric state. Although temperate viruses are thought to share a similar lytic-lysogenic cycle switch model, our study suggests the existence of alternative strain-specific mechanisms that regulate the lysis-lysogeny decision. Collectively, these findings provide insights into the molecular mechanisms underlying communication among viruses, offering theoretical applications for the treatment of infectious viral diseases.
Organizational Affiliation:
Division of Respiratory and Critical Care Medicine, State Key Laboratory of Biotherapy, West China Hospital of Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu, China.