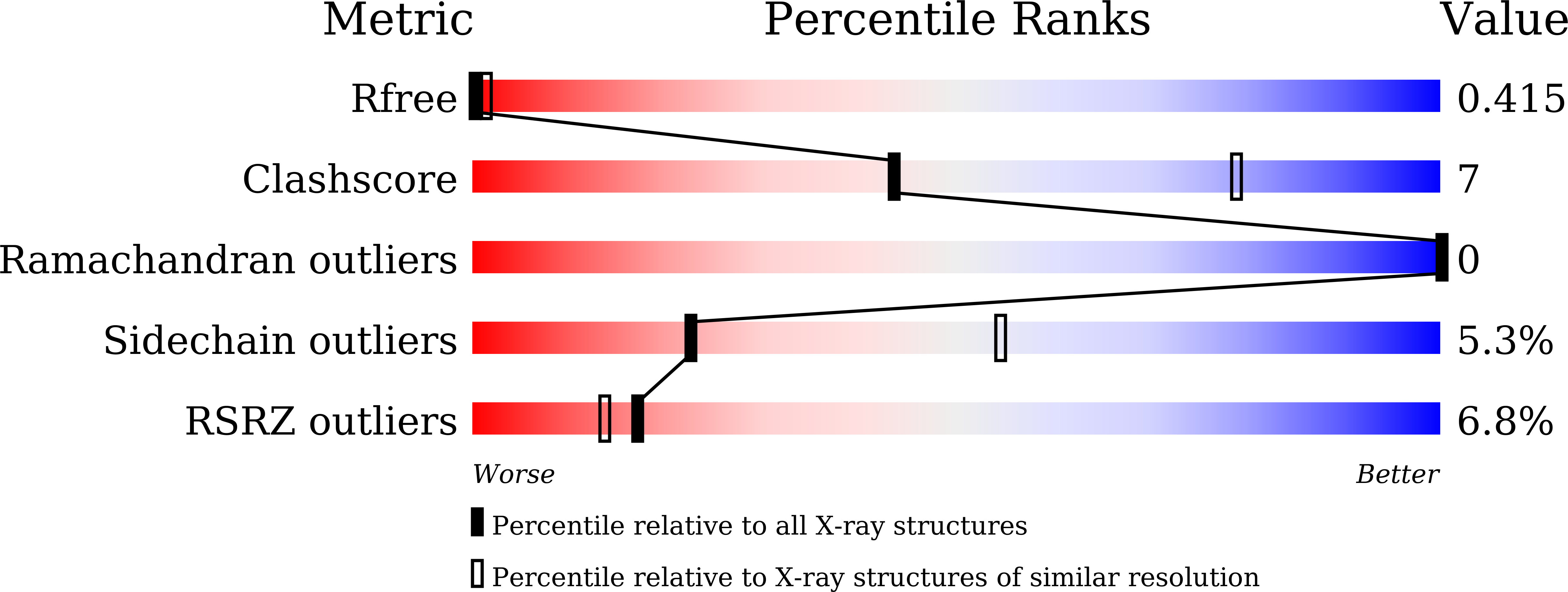
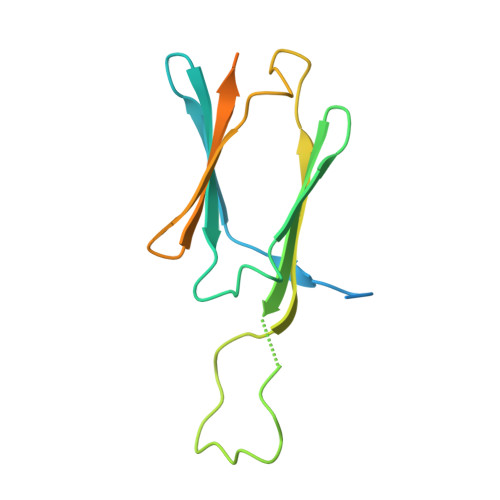
Dodecameric structure of a small heat shock protein from Mycobacterium marinum M.
Bhandari, S., Biswas, S., Chaudhary, A., Dutta, S., Suguna, K.(2019) Proteins 87: 365-379
- PubMed: 30632633
- DOI: https://doi.org/10.1002/prot.25657
- Primary Citation of Related Structures:
5ZS3, 5ZS6, 5ZUL - PubMed Abstract:
Small heat shock proteins (sHSPs) are ATP-independent molecular chaperones present ubiquitously in all kingdoms of life. Their low molecular weight subunits associate to form higher order structures. Under conditions of stress, sHSPs prevent aggregation of substrate proteins by undergoing rapid changes in their conformation or stoichiometry. Polydispersity and dynamic nature of these proteins have made structural investigations through crystallography a daunting task. In pathogens like Mycobacteria, sHSPs are immuno-dominant antigens, enabling survival of the pathogen within the host and contributing to disease persistence. We characterized sHSPs from Mycobacterium marinum M and determined the crystal structure of one of these. The protein crystallized in three different conditions as dodecamers, with dimers arranged in a tetrahedral fashion to form a closed cage-like architecture. Interestingly, we found a pentapeptide bound to the dodecamers revealing one of the modes of sHSP-substrate interaction. Further, we have observed that ATP inhibits the chaperoning activity of the protein.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.